1FXF
 
 | | CARBOXYLIC ESTER HYDROLASE COMPLEX (DIMERIC PLA2 + MJ33 INHIBITOR + PHOSPHATE IONS) | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Pan, Y.H, Epstein, T.M, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Five coplanar anion binding sites on one face of phospholipase A2: relationship to interface binding.
Biochemistry, 40, 2001
|
|
6PWH
 
 | |
7N6D
 
 | | HLA peptide complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Chaurasia, P, Petersen, J, Rossjohn, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein.
J.Biol.Chem., 297, 2021
|
|
4DRW
 
 | | Crystal Structure of the Ternary Complex between S100A10, an Annexin A2 N-terminal Peptide and an AHNAK Peptide | | Descriptor: | Neuroblast differentiation-associated protein AHNAK, Protein S100-A10/Annexin A2 chimeric protein | | Authors: | Rezvanpour, A, Lee, T.-W, Junop, M.S, Shaw, G.S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of an asymmetric ternary protein complex provides insight for membrane interaction.
Structure, 20, 2012
|
|
8DWM
 
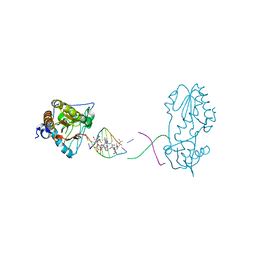 | | Host-guest complex of bleomycin A2 fully bound to CTTAGTTATAACTAAG | | Descriptor: | BLEOMYCIN A2, COBALT (III) ION, DNA (5'-D(*CP*TP*TP*AP*GP*TP*TP*A)-3'), ... | | Authors: | Georgiadis, M.M. | | Deposit date: | 2022-08-01 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Two distinct rotations of bithiazole DNA intercalation revealed by direct comparison of crystal structures of Co(III)•bleomycin A 2 and B 2 bound to duplex 5'-TAGTT sites.
Bioorg.Med.Chem., 77, 2023
|
|
2WG9
 
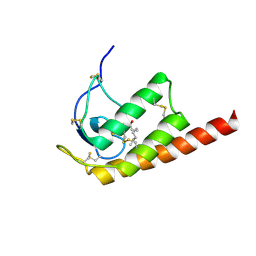 | | Structure of Oryza Sativa (Rice) PLA2, complex with octanoic acid | | Descriptor: | CALCIUM ION, OCTANOIC ACID (CAPRYLIC ACID), PUTATIVE PHOSPHOLIPASE A2, ... | | Authors: | Guy, J.E, Stahl, U, Lindqvist, Y. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Class Xib Phospholipase A2 (Pla2): Rice (Oryza Sativa) Isoform-2 Pla2 and an Octanoate Complex.
J.Biol.Chem., 284, 2009
|
|
1GP7
 
 | |
2WG8
 
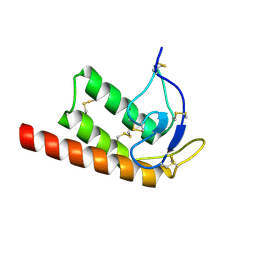 | | Structure of Oryza Sativa (Rice) PLA2, orthorhombic crystal form | | Descriptor: | CALCIUM ION, PUTATIVE PHOSPHOLIPASE A2, SODIUM ION | | Authors: | Guy, J.E, Stahl, U, Lindqvist, Y. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Class Xib Phospholipase A2 (Pla2): Rice (Oryza Sativa) Isoform-2 Pla2 and an Octanoate Complex.
J.Biol.Chem., 284, 2009
|
|
2WG7
 
 | | Structure of Oryza Sativa (Rice) PLA2 | | Descriptor: | CALCIUM ION, PUTATIVE PHOSPHOLIPASE A2, SODIUM ION | | Authors: | Guy, J.E, Stahl, U, Lindqvist, Y. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Class Xib Phospholipase A2 (Pla2): Rice (Oryza Sativa) Isoform-2 Pla2 and an Octanoate Complex.
J.Biol.Chem., 284, 2009
|
|
5YD6
 
 | | Crystal structure of PG-bound Nurr1-LBD | | Descriptor: | (~{Z})-7-[(1~{R},5~{S})-2-oxidanylidene-5-[(~{E},3~{S})-3-oxidanyloct-1-enyl]cyclopent-3-en-1-yl]hept-5-enoic acid, MAGNESIUM ION, Nuclear receptor subfamily 4 group A member 2 | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2017-09-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of Nurr1 bound to cyclopentenone prostaglandin A2 and its mechanism of action in ameliorating dopaminergic neurodegeneration in Drosophila
To Be Published
|
|
5YBQ
 
 | |
3HHV
 
 | | The crystal structure of the Thioredoxin A2 from Sulfolobus solfataricus | | Descriptor: | Thioredoxin (TrxA-2) | | Authors: | Ruggiero, A, Masullo, M, Ruocco, M.R, Arcari, P, Zagari, A, Vitagliano, L. | | Deposit date: | 2009-05-18 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The dimeric structure of Sulfolobus solfataricus thioredoxin A2 and the basis of its thermostability
Proteins, 77, 2009
|
|
5E6I
 
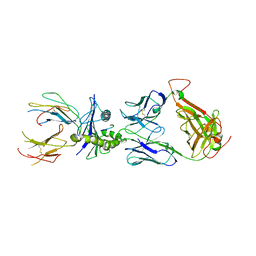 | |
9C4A
 
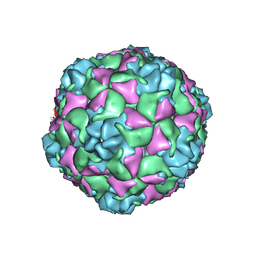 | | Cryo-EM Structure of EV-D68 Vaccine Candidate - A2 Subclade Virus-like Particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Cheng, J, Krug, P.W, Lei, H, Moss, D.L, Huang, R, Lang, Z.C, Morton, A.J, Shen, C, Pierson, T.C, Zhou, T, Ruckwardt, T.J, Kwong, P.D. | | Deposit date: | 2024-06-03 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Cryo-EM Structure of EV-D68 Vaccine Candidate - A2 Subclade Virus-like Particle
To Be Published
|
|
5EUO
 
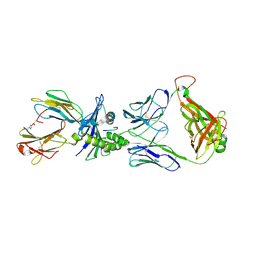 | | PF6-M1-HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Yang, X. | | Deposit date: | 2015-11-18 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PF6-M1-HLA-A2 complex
To Be Published
|
|
4FA0
 
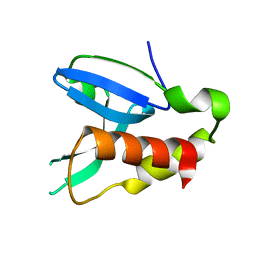 | | Crystal structure of human AdPLA to 2.65 A resolution | | Descriptor: | Group XVI phospholipase A1/A2 | | Authors: | Lovell, S, Battaile, K.P, Addington, L, Zhang, N, Rao, J.L.U.M, Moise, A.R. | | Deposit date: | 2012-05-21 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure/Function relationships of adipose phospholipase A2 containing a cys-his-his catalytic triad.
J.Biol.Chem., 287, 2012
|
|
5NHT
 
 | | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
3AL8
 
 | | Plexin A2 / Semaphorin 6A complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A2, ... | | Authors: | Nogi, T, Yasui, N, Mihara, E, Takagi, J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
3AL9
 
 | | Mouse Plexin A2 extracellular domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A2 | | Authors: | Nogi, T, Yasui, N, Mihara, E, Takagi, J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for semaphorin signalling through the plexin receptor.
Nature, 467, 2010
|
|
2ZW7
 
 | |
7AYX
 
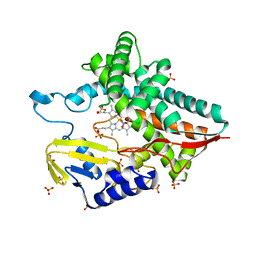 | |
5NQK
 
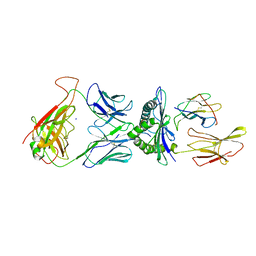 | | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-04-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
7D5S
 
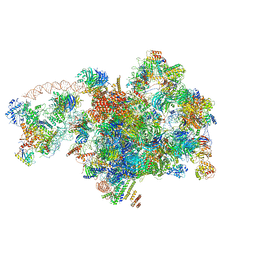 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S12, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state A2)
To Be Published
|
|
4HX1
 
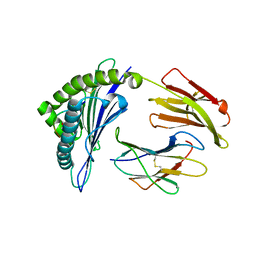 | | Structure of HLA-A68 complexed with a tumor antigen derived peptide | | Descriptor: | 9-mer peptide from Tyrosinase-related protein-2, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-09 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4I48
 
 | | Structure of HLA-A68 complexed with an HIV Env derived peptide | | Descriptor: | 9-mer peptide from Envelope glycoprotein gp160, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-27 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
