7LRO
 
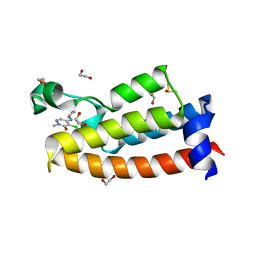 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
4QUZ
 
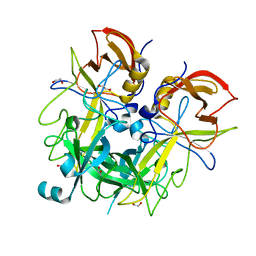 | |
6A03
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6YOC
 
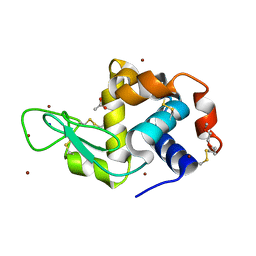 | | Structure of Lysozyme from COC IMISX setup collected by still serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
7ZT7
 
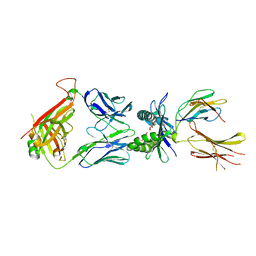 | |
8QYR
 
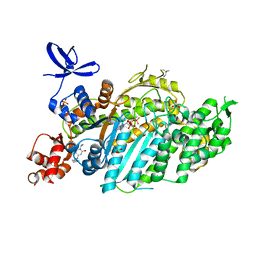 | | Beta-cardiac myosin motor domain in the pre-powerstroke state complexed to Mavacamten | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Robert-Paganin, J, Kikuti, C, Auguin, D, Rety, S, David, A, Houdusse, A. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-13 | | Last modified: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite antagonistic effects in heart contraction.
Biorxiv, 2023
|
|
8EUD
 
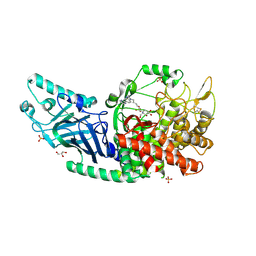 | | Co-crystal structure of Chaetomium glucosidase with compound 22 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{6-[2-methyl-5-(pyrimidin-2-yl)-1H-benzimidazol-1-yl]hexyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6YPE
 
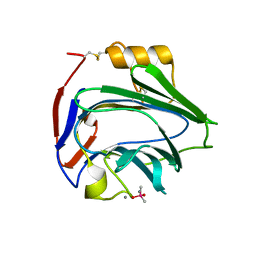 | |
8CFH
 
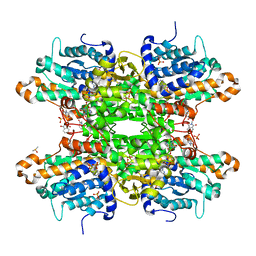 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment C08 | | Descriptor: | (3S)-3-hydroxy-2-methyl-2,3-dihydro-1H-isoindol-1-one, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry C08
To be published
|
|
8W3W
 
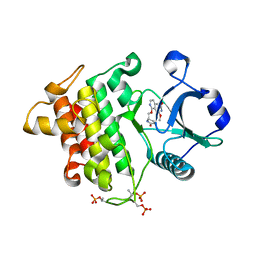 | | Crystal structure of IRAK4 in complex with compound 4 | | Descriptor: | 7-methoxy-1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | In Retrospect: Root-Cause Analysis of Structure-Activity Relationships in IRAK4 Inhibitor Zimlovisertib (PF-06650833).
Acs Med.Chem.Lett., 15, 2024
|
|
5MOE
 
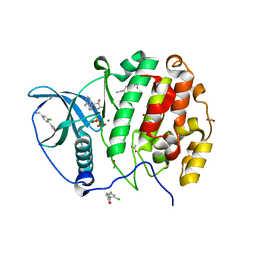 | | Crystal Structure of CK2alpha with N-(3-(((2-chloro-[1,1'-biphenyl]-4-yl)methyl)amino)propyl)methanesulfonamide bound | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
8CFR
 
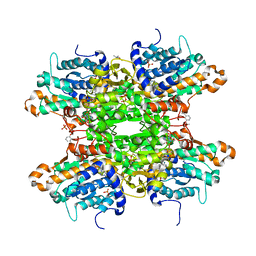 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment G03 | | Descriptor: | 3-ethoxybenzene-1-carboximidamide, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry G03
To be published
|
|
8W3X
 
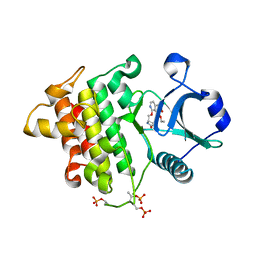 | | Crystal structure of IRAK4 in complex with compound 6 | | Descriptor: | 7-ethoxy-1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | In Retrospect: Root-Cause Analysis of Structure-Activity Relationships in IRAK4 Inhibitor Zimlovisertib (PF-06650833).
Acs Med.Chem.Lett., 15, 2024
|
|
7LP4
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
8SMC
 
 | | Cryo-EM structure of LRRK2 bound with type-I inhibitor DNL201 | | Descriptor: | 2-methyl-2-(3-methyl-4-{[4-(methylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}-1H-pyrazol-1-yl)propanenitrile, GUANOSINE-5'-DIPHOSPHATE, non-specific serine/threonine protein kinase | | Authors: | Sun, J, Zhu, H. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Rab29-dependent asymmetrical activation of leucine-rich repeat kinase 2.
Science, 382, 2023
|
|
8CFY
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment H08 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry H08
To be published
|
|
7QTW
 
 | | Kaposi sarcoma associated herpes virus(KSHV) encoded apoptosis inhibitor, KsBcl-2 in complex with Bid BH3 | | Descriptor: | 1,2-ETHANEDIOL, BH3-interacting domain death agonist p15, Bcl-2 | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Insight into KsBcl-2 Mediated Apoptosis Inhibition by Kaposi Sarcoma Associated Herpes Virus.
Viruses, 14, 2022
|
|
7ZLU
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-2-deoxy-2-fluoro-D-glucose | | Descriptor: | 1,3-PROPANDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Ibba, R. | | Deposit date: | 2022-04-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
8K3U
 
 | | S. cerevisiae Chs1 in complex with UDP and GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin synthase 1, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
6Y34
 
 | | Streptavidin wildtype with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.307 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
8EPO
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 18 | | Descriptor: | (3P)-3-(5,6-dihydro-1,4-dioxin-2-yl)-5-{[(3-{[(2R,3R,4R,5S)-3,4,5-trihydroxy-2-(hydroxymethyl)piperidin-1-yl]methyl}phenyl)methyl]amino}benzonitrile, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8CFB
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment D02 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment D02
To be published
|
|
8K3R
 
 | | S. cerevisiae Chs1 in apo state incubated with GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8CLQ
 
 | |
8CFO
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment F04 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry F04
To be published
|
|
