1N8N
 
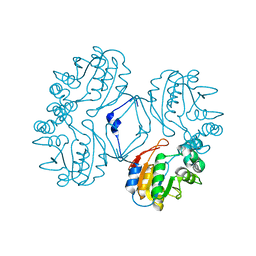 | | Crystal structure of the Au3+ complex of AphA class B acid phosphatase/phosphotransferase from E. coli at 1.69 A resolution | | Descriptor: | Class B acid phosphatase, GOLD 3+ ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2002-11-21 | | Release date: | 2004-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The first structure of a bacterial class B Acid phosphatase reveals further structural heterogeneity among phosphatases of the haloacid dehalogenase fold.
J.Mol.Biol., 335, 2004
|
|
1RI3
 
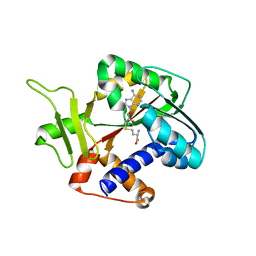 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RJX
 
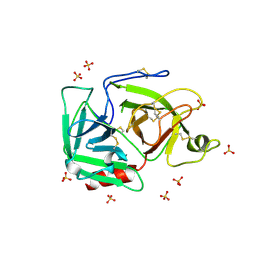 | | Human PLASMINOGEN CATALYTIC DOMAIN, K698M MUTANT | | Descriptor: | Plasminogen, SULFATE ION | | Authors: | Terzyan, S, Wakeham, N, Zhai, P, Rodgers, K, Zhang, X.C. | | Deposit date: | 2003-11-20 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Lys-698 to met substitution in human plasminogen catalytic domain
Proteins, 56, 2004
|
|
1R1U
 
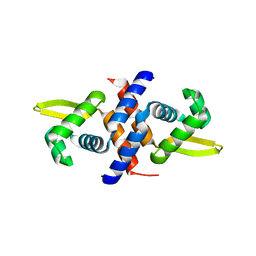 | | Crystal structure of the metal-sensing transcriptional repressor CzrA from Staphylococcus aureus in the apo-form | | Descriptor: | repressor protein | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R23
 
 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB in the Zn1-form (one Zn(II) per dimer) | | Descriptor: | Transcriptional repressor smtB, ZINC ION | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
2CIO
 
 | |
1R6K
 
 | | HPV11 E2 TAD crystal structure | | Descriptor: | HPV11 REGULATORY PROTEIN E2 | | Authors: | Wang, Y, Coulombe, R. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the E2 Transactivation Domain of Human Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
1R74
 
 | | Crystal Structure of Human Glycine N-Methyltransferase | | Descriptor: | BETA-MERCAPTOETHANOL, CITRIC ACID, Glycine N-methyltransferase | | Authors: | Pakhomova, S, Luka, Z, Wagner, C, Newcomer, M.E. | | Deposit date: | 2003-10-17 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes.
Proteins, 57, 2004
|
|
1R7W
 
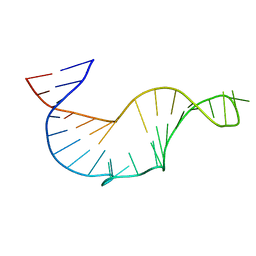 | | NMR STRUCTURE OF THE R(GGAGGACAUCCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUCCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1R3D
 
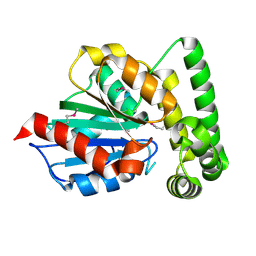 | |
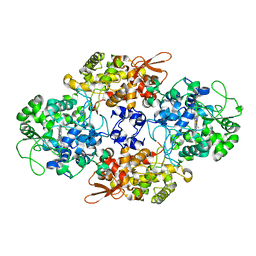
2CCD
 
 | |
1RF6
 
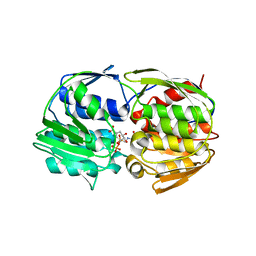 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in S3P-GLP Bound State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase, GLYPHOSATE, SHIKIMATE-3-PHOSPHATE | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
1PSN
 
 | | THE CRYSTAL STRUCTURE OF HUMAN PEPSIN AND ITS COMPLEX WITH PEPSTATIN | | Descriptor: | PEPSIN 3A | | Authors: | Fujinaga, M, Chernaia, M.M, Tarasova, N, Mosimann, S.C, James, M.N.G. | | Deposit date: | 1995-01-23 | | Release date: | 1995-04-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human pepsin and its complex with pepstatin.
Protein Sci., 4, 1995
|
|
2AZT
 
 | | Crystal structure of H176N mutant of human Glycine N-Methyltransferase | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Luka, Z, Pakhomova, S, Luka, Y, Newcomer, M.E, Wagner, C. | | Deposit date: | 2005-09-12 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Destabilization of human glycine N-methyltransferase by H176N mutation.
Protein Sci., 16, 2007
|
|
1RMT
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase complexed with adenosine. | | Descriptor: | ADENOSINE, Class B acid phosphatase, MAGNESIUM ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
1Q8P
 
 | | Pterocarpus angolensis lectin PAL in complex with the dimannoside Man(alpha1-3)Man | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, alpha-D-mannopyranose-(1-3)-methyl alpha-D-mannopyranoside, ... | | Authors: | Loris, R, Van Walle, I, De Greve, H, Beeckmans, S, Deboeck, F, Wyns, L, Bouckaert, J. | | Deposit date: | 2003-08-22 | | Release date: | 2004-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Oligomannose Recognition by the Pterocarpus angolensis Seed Lectin
J.Mol.Biol., 335, 2004
|
|
2B4R
 
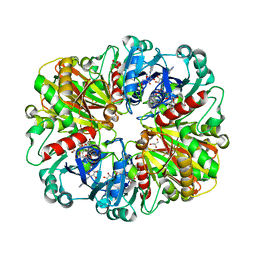 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 Angstrom Resolution reveals intriguing extra electron density in the active site | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Robien, M.A, Bosch, J, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site
Proteins, 62, 2006
|
|
1R7Z
 
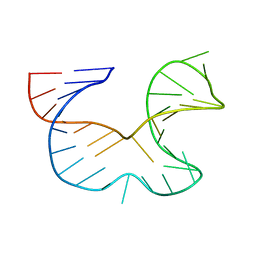 | | NMR STRUCTURE OF THE R(GGAGGACAUUCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUUCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1V4E
 
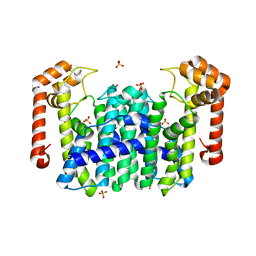 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
1UY7
 
 | | Human Hsp90-alpha with 9-Butyl-8-(4-methoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(4-METHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
2AM9
 
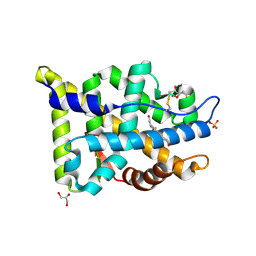 | | Crystal structure of human androgen receptor ligand binding domain in complex with testosterone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Androgen receptor, GLYCEROL, ... | | Authors: | Pereira de Jesus-Tran, K, Cote, P.-L, Cantin, L, Blanchet, J, Labrie, F, Breton, R. | | Deposit date: | 2005-08-09 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity.
Protein Sci., 15, 2006
|
|
2B3T
 
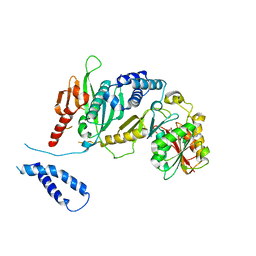 | | Structure of complex between E. coli translation termination factor RF1 and the PrmC methyltransferase | | Descriptor: | Peptide chain release factor 1, Protein methyltransferase hemK, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Graille, M, Heurgue-Hamard, V, Champ, S, Mora, L, Scrima, N, Ulryck, N, van Tilbeurgh, H, Buckingham, R.H. | | Deposit date: | 2005-09-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis for bacterial class I release factor methylation by PrmC
Mol.Cell, 20, 2005
|
|
1QVZ
 
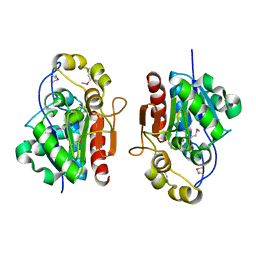 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1V38
 
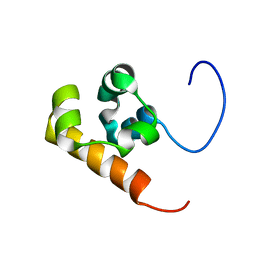 | | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1 | | Descriptor: | SAM-domain protein SAMSN-1 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-29 | | Release date: | 2004-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1
To be Published
|
|
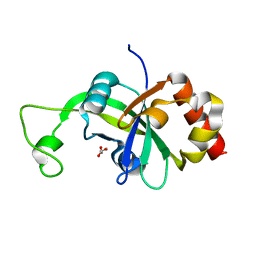
2AR1
 
 | |
