9QEG
 
 | | Cryo-EM structure of the 70S ribosome of a MLSb sensitive S. aureus strain "KES34" in complex with solithromycin | | Descriptor: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Rivalta, A, Yonath, A. | | Deposit date: | 2025-03-10 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural studies on ribosomes of differentially macrolide-resistant Staphylococcus aureus strains.
Life Sci Alliance, 8, 2025
|
|
6XS9
 
 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-L1 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 48V-TYR-ILE-LYS-THR-PRO-LEU-GLY-THR-PHE-PRO-ASN-ARG-HIS-GLY, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
8C3V
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-13 Fab and C1 nanobody | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, BA.2-13 heavy chain, BA.2-13 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-12-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
6NJZ
 
 | |
8QNU
 
 | |
7AD6
 
 | | Crystal structure of human complement C5 in complex with the K92 bovine knob domain peptide. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYSTEINE, ... | | Authors: | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
7PGJ
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-decarboxylate TamK and 10-hydroxylase RdmB, together with a single point mutation F297G | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-7-methoxy-2,4,5-tris(oxidanyl)-6,11-bis(oxidanylidene)-3,4-dihydro-1H-tetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
6PCS
 
 | | E. coli 50S ribosome bound to compound 40e | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl [4-(trifluoromethyl)phenyl]carbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
8QM9
 
 | | Potential drug binding sites for translation initiation factor eIF4E | | Descriptor: | (2~{R})-2-[(1~{S})-1-[4-(2-fluorophenyl)-2-(2-hydroxyethylamino)phenyl]propoxy]propan-1-ol, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cleasby, A. | | Deposit date: | 2023-09-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Integrating fragment-based screening with targeted protein degradation and genetic rescue to explore eIF4E function.
Nat Commun, 15, 2024
|
|
6PJ0
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-5 (NR01-97) | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4 Aprotease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
7PYH
 
 | | Structure of LPMO in complex with cellotetraose at 1.45x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYF
 
 | | Structure of LPMO in complex with cellotetraose at 1.39x10^5 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
8HFY
 
 | | SARS-CoV-2 Omicron BA.1 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8AGZ
 
 | | Yeast RQC complex in state with the RING domain of Ltn1 in the OUT position | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
6P0M
 
 | | Crystal structure of GDP-bound human RalA in a covalent complex with aryl sulfonyl fluoride compounds. | | Descriptor: | 4-[(3-chloropyridin-2-yl)sulfamoyl]benzene-1-sulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Ghozayel, M.K, Xu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7NVR
 
 | | Human Mediator with RNA Polymerase II Pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Rengachari, S, Schilbach, S, Aibara, S, Cramer, P. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7PYU
 
 | | Structure of an LPMO (expressed in E.coli) at 1.49x10^4 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7Q1K
 
 | | Crystal structure of the native AA9A LPMO from Thermoascus aurantiacus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Yu, W, Mohsin, I, Li, D.C, Papageorgiou, A.C. | | Deposit date: | 2021-10-20 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Purification and Structural Characterization of the Auxiliary Activity 9 Native Lytic Polysaccharide Monooxygenase from Thermoascus aurantiacus and Identification of Its C1- and C4-Oxidized Reaction Products
Catalysts, 12, 2022
|
|
6XRN
 
 | | Crystal structure of human PI3K-gamma in complex with Compound 17 | | Descriptor: | 2-amino-5-{2-[(1S)-1-cyclopropylethyl]-7-methyl-1-oxo-2,3-dihydro-1H-isoindol-5-yl}-N-(trans-3-hydroxycyclobutyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationship Optimization of Pyrazolopyrimidine Amide Inhibitors of Phosphoinositide 3-Kinase gamma (PI3K gamma ).
J.Med.Chem., 65, 2022
|
|
6P12
 
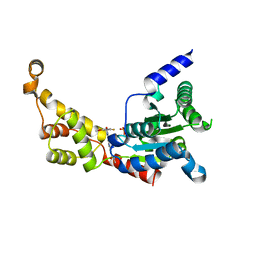 | | Structure of spastin AAA domain (wild-type) in complex with diaminotriazole-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, Drosophila melanogaster Spastin AAA domain, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94131851 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
8FKY
 
 | |
8FKV
 
 | |
8FKU
 
 | |
8FKX
 
 | |
8FKS
 
 | |
