2LRR
 
 | | Solution structure of the R3H domain from human Smubp-2 in complex with 2'-deoxyguanosine-5'-monophosphate | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-binding protein SMUBP-2 | | Authors: | Jaudzems, K, Zhulenkovs, D, Otting, G, Liepinsh, E. | | Deposit date: | 2012-04-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for 5'-End-Specific Recognition of Single-Stranded DNA by the R3H Domain from Human Smubp-2
J.Mol.Biol., 12, 2012
|
|
2LKS
 
 | | Ff11-60 | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Barette, J, Velyvis, A, Religa, T.L, Korzhnev, D.M, Kay, L.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cross-Validation of the Structure of a Transiently Formed and Low Populated FF Domain Folding Intermediate Determined by Relaxation Dispersion NMR and CS-Rosetta.
J.Phys.Chem.B, 116, 2012
|
|
3UBR
 
 | | Laue structure of Shewanella oneidensis cytochrome-c Nitrite Reductase | | Descriptor: | CALCIUM ION, Cytochrome c-552, HEME C | | Authors: | Youngblut, M, Judd, E.T, Srajer, V, Sayed, B, Goeltzner, T, Elliott, S, Schmidt, M, Pacheco, A. | | Deposit date: | 2011-10-24 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system.
J.Biol.Inorg.Chem., 17, 2012
|
|
3TMV
 
 | | X-Ray Radiation Damage to HEWL Crystals soaked in 100mM Sodium Nitrate (Dose=0.12MGy) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Kmetko, J, Warkentin, M.A, Englich, U, Thorne, R.E. | | Deposit date: | 2011-08-31 | | Release date: | 2012-08-22 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Can radiation damage to protein crystals be reduced using small-molecule compounds?
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2LGT
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for QFM(Y)F | | Descriptor: | Eukaryotic peptide chain release factor subunit 1 | | Authors: | Wong, L.E, Li, Y, Pillay, S, Pervushin, K. | | Deposit date: | 2011-08-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Selectivity of stop codon recognition in translation termination is modulated by multiple conformations of GTS loop in eRF1
Nucleic Acids Res., 2012
|
|
2LD0
 
 | |
4ZCR
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase in complex with phosphocholine | | Descriptor: | Cholinephosphate cytidylyltransferase, PHOSPHOCHOLINE | | Authors: | Guca, E, Hoh, F, Guichou, J.-F, Cerdan, R. | | Deposit date: | 2015-04-16 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of the catalytic mechanism of Plasmodium CCT, a key enzyme of malaria lipid biosynthesis.
Sci Rep, 8, 2018
|
|
3E4B
 
 | | Crystal structure of AlgK from Pseudomonas fluorescens WCS374r | | Descriptor: | AlgK, CHLORIDE ION, GLYCEROL | | Authors: | Keiski, C.-L, Harwich, M, Jain, S, Neculai, A.M, Yip, P, Robinson, H, Whitney, J.C, Burrows, L.L, Ohman, D.E, Howell, P.L. | | Deposit date: | 2008-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin.
Structure, 18, 2010
|
|
4ZCT
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase | | Descriptor: | Cholinephosphate cytidylyltransferase | | Authors: | Guca, E, Hoh, F, Guichou, J.-F, Cerdan, R. | | Deposit date: | 2015-04-16 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural determinants of the catalytic mechanism of Plasmodium CCT, a key enzyme of malaria lipid biosynthesis.
Sci Rep, 8, 2018
|
|
3F5V
 
 | | C2 Crystal form of mite allergen DER P 1 | | Descriptor: | CALCIUM ION, Der p 1 allergen, HEXAETHYLENE GLYCOL | | Authors: | Stura, E.A, Minor, W, Chruszcz, M, Saint Remy, J.M. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures of mite allergens Der f 1 and Der p 1 reveal differences in surface-exposed residues that may influence antibody binding.
J.Mol.Biol., 386, 2009
|
|
3FE9
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera with a serendipitous ligand soaked at pH 7.0 | | Descriptor: | (20S)-20-methyldotetracontane, CHLORIDE ION, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Queen bee pheromone binding protein pH induced domain-swapping favors pheromone release
To be Published
|
|
5A24
 
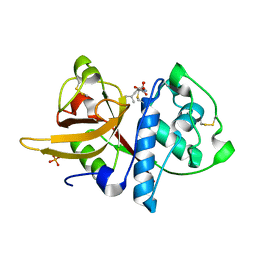 | | Crystal structure of Dionain-1, the major endopeptidase in the Venus flytrap digestive juice | | Descriptor: | DIONAIN-1, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, PHOSPHATE ION | | Authors: | Risor, M.W, Thomsen, L.R, Sanggaard, K.W, Nielsen, T.A, Thogersen, I.B, Lukassen, M.V, Rossen, L, Garcia-Ferrer, I, Guevara, T, Meinjohanns, E, Nielsen, N.C, Gomis-Ruth, F.X, Enghild, J.J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enzymatic and Structural Characterization of the Major Endopeptidase in the Venus Flytrap Digestion Fluid.
J.Biol.Chem., 291, 2016
|
|
4ZCQ
 
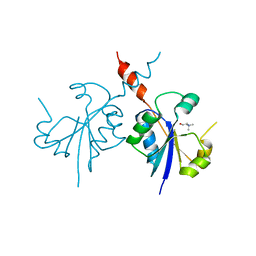 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase in complex with choline | | Descriptor: | CHOLINE ION, Cholinephosphate cytidylyltransferase | | Authors: | Guca, E, Hoh, F, Guichou, J.-F, Cerdan, R. | | Deposit date: | 2015-04-16 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural determinants of the catalytic mechanism of Plasmodium CCT, a key enzyme of malaria lipid biosynthesis.
Sci Rep, 8, 2018
|
|
4ZCP
 
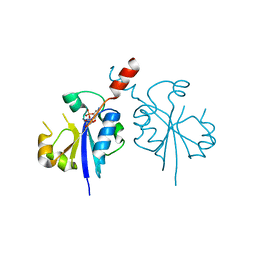 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase in complex with CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Cholinephosphate cytidylyltransferase | | Authors: | Guca, E, Hoh, F, Guichou, J.-F, Cerdan, R. | | Deposit date: | 2015-04-16 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural determinants of the catalytic mechanism of Plasmodium CCT, a key enzyme of malaria lipid biosynthesis.
Sci Rep, 8, 2018
|
|
1Z99
 
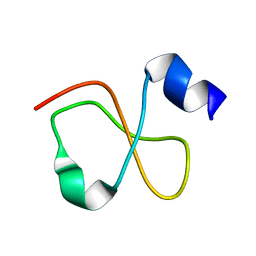 | | Solution structure of Crotamine, a myotoxin from Crotalus durissus terrificus | | Descriptor: | Crotamine | | Authors: | Fadel, V, Bettendorff, P, Herrmann, T, de Azevedo, W.F, Oliveira, E.B, Yamane, T, Wuthrich, K. | | Deposit date: | 2005-04-01 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Automated NMR structure determination and disulfide bond identification of the myotoxin crotamine from Crotalus durissus terrificus.
Toxicon, 46, 2005
|
|
1YOD
 
 | |
3G1A
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3EUN
 
 | |
3G1X
 
 | | Crystal structure of the mutant D70G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3G1D
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3G22
 
 | | Crystal structure of the mutant D70N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3G1V
 
 | | Crystal structure of the mutant D70G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 5-fluorouridine 5'-monophosphate | | Descriptor: | 5-FLUORO-URIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3G24
 
 | | Crystal structure of the mutant D70N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3GDL
 
 | | Crystal structure of the orotidine 5'-monophosphate decarboxylase from Saccharomyces cerevisiae complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3EXY
 
 | |
