3MAS
 
 | |
3MAB
 
 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED PROTEIN FROM LISTERIA MONOCYTOGENES, Triclinic FORM | | Descriptor: | uncharacterized protein | | Authors: | Madegowda, M, Chruszcz, M, Minor, W, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of an uncharacterized protein from listeria monocytogenes
To be Published
|
|
3MBP
 
 | | MALTODEXTRIN-BINDING PROTEIN WITH BOUND MALTOTRIOSE | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Spurlino, J.C, Quiocho, F.A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor.
Structure, 5, 1997
|
|
3W9T
 
 | | pore-forming CEL-III | | Descriptor: | CALCIUM ION, Hemolytic lectin CEL-III, MAGNESIUM ION, ... | | Authors: | Unno, H, Goda, S, Hatakeyama, T. | | Deposit date: | 2013-04-16 | | Release date: | 2014-03-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hemolytic lectin CEL-III heptamer reveals its transmembrane pore-formation mechanism
J.Biol.Chem., 2014
|
|
3M78
 
 | |
3MCN
 
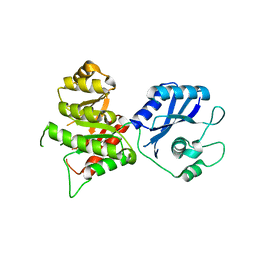 | | Crystal Structure of the 6-hyroxymethyl-7,8-dihydropterin pyrophosphokinase dihydropteroate synthase bifunctional enzyme from Francisella tularensis | | Descriptor: | 2,6-diamino-5-nitropyrimidin-4(3H)-one, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, MAGNESIUM ION | | Authors: | Pemble IV, C.W, Mehta, P.K, Mehra, S, Li, Z, Lee, R.E, White, S.W. | | Deposit date: | 2010-03-29 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase.dihydropteroate synthase bifunctional enzyme from Francisella tularensis.
Plos One, 5, 2010
|
|
3MDZ
 
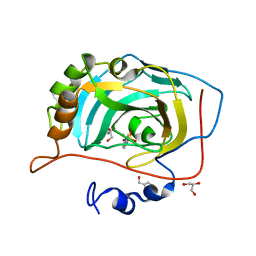 | | Crystal Structure of Human Carbonic Anhydrase VII [isoform 1], CA7 | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 7, GLYCEROL, ... | | Authors: | Ugochukwu, E, Shafqat, N, Pilka, E, Chaikuad, A, Krojer, T, Muniz, J, Kim, J, Bray, J, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Carpenter, E.P, Yue, W.W, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of Human Carbonic Anhydrase VII [isoform 1], CA7
to be published
|
|
3WSE
 
 | | Reduced HcgD from Methanocaldococcus jannaschii | | Descriptor: | FE (II) ION, PHOSPHATE ION, Putative GTP cyclohydrolase 1 type 2 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A possible iron delivery function of the dinuclear iron center of HcgD in [Fe]-hydrogenase cofactor biosynthesis
Febs Lett., 588, 2014
|
|
3MDY
 
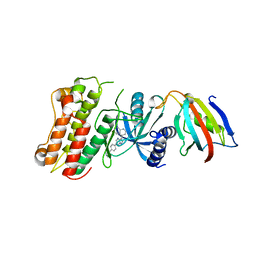 | | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Bone morphogenetic protein receptor type-1B, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chaikuad, A, Sanvitale, C, Mahajan, P, Daga, N, Cooper, C, Krojer, T, Alfano, I, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189
To be Published
|
|
3MJ9
 
 | |
3WUS
 
 | | Crystal Structure of the Vif-Binding Domain of Human APOBEC3F | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3F, ZINC ION | | Authors: | Nakashima, M, Kawamura, T, Ode, H, Watanabe, N, Iwatani, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Insights into HIV-1 Vif-APOBEC3F Interaction.
J.Virol., 90, 2015
|
|
3MM7
 
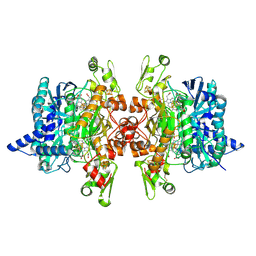 | | Dissimilatory sulfite reductase carbon monoxide complex | | Descriptor: | CARBON MONOXIDE, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3W7V
 
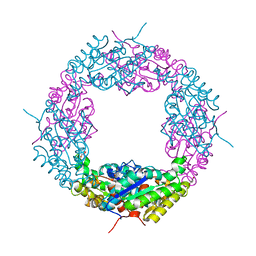 | | Crystal Structure of Axe2, an Acetylxylan Esterase from Geobacillus stearothermophilus | | Descriptor: | Acetyl xylan esterase, CHLORIDE ION, GLYCEROL | | Authors: | Lansky, S, Alalouf, O, Solomon, H.V, Alhassid, A, Belrahli, H, Govada, L, Chayan, N.E, Shoham, Y, Shoham, G. | | Deposit date: | 2013-03-08 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A unique octameric structure of Axe2, an intracellular acetyl-xylooligosaccharide esterase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3MOD
 
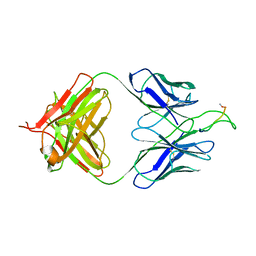 | | Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced IgG) with 11 aa gp41 MPER-derived peptide | | Descriptor: | ANTI-HIV-1 ANTIBODY 2F5 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 2F5 LIGHT CHAIN, gp41 MPER-derived peptide | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
3W8X
 
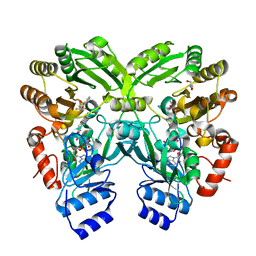 | | The complex structure of EncM with trifluorotriketide | | Descriptor: | 6,6,6-trifluoro-1-phenylhexane-1,3,5-trione, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3WBN
 
 | | Crystal structure of MATE in complex with MaL6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MaL6, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3W8Z
 
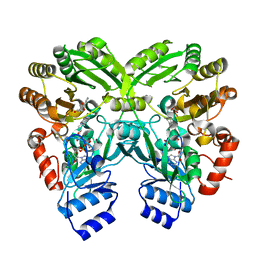 | | The complex structure of EncM with hydroxytetraketide | | Descriptor: | (7S)-7-hydroxy-1-phenyloctane-1,3,5-trione, FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3WIW
 
 | | Crystal structure of unsaturated glucuronyl hydrolase specific for heparin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycosyl hydrolase family 88 | | Authors: | Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a bacterial unsaturated glucuronyl hydrolase with specificity for heparin.
J.Biol.Chem., 289, 2014
|
|
3M6E
 
 | |
3M72
 
 | |
3M7N
 
 | |
3WBO
 
 | |
3MCM
 
 | | Crystal Structure of the 6-hyroxymethyl-7,8-dihydropterin pyrophosphokinase dihydropteroate synthase bifunctional enzyme from Francisella tularensis | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, MAGNESIUM ION | | Authors: | Pemble IV, C.W, Mehta, P.K, Mehra, S, Li, Z, Lee, R.E, White, S.W. | | Deposit date: | 2010-03-29 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase.dihydropteroate synthase bifunctional enzyme from Francisella tularensis.
Plos One, 5, 2010
|
|
3M74
 
 | |
3WRU
 
 | | Crystal structure of the bacterial ribosomal decoding site in complex with synthetic aminoglycoside with F-HABA group | | Descriptor: | (2R,3R)-4-amino-N-[(1R,2S,3R,4R,5S)-5-amino-4-[(2,6-diamino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranosyl)oxy]-3-{[3-O-(2,6-diamino-2,3,4,6-tetradeoxy-beta-L-threo-hexopyranosyl)-beta-D-ribofuranosyl]oxy}-2-hydroxycyclohexyl]-3-fluoro-2-hydroxybutanamide, POTASSIUM ION, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J, Hanessian, S. | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Toxicity Modulation, Resistance Enzyme Evasion, and A-Site X-ray Structure of Broad-Spectrum Antibacterial Neomycin Analogs
Acs Chem.Biol., 9, 2014
|
|
