2C65
 
 | | MAO inhibition by rasagiline analogues | | Descriptor: | (1R)-4-({[ETHYL(METHYL)AMINO]CARBONYL}OXY)-N-METHYL-N-[(1E)-PROP-2-EN-1-YLIDENE]INDAN-1-AMINIUM, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-11-07 | | Release date: | 2006-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of Rasagiline-Related Inhibitors to Human Monoamine Oxidases: A Kinetic and Crystallographic Analysis.
J.Med.Chem., 48, 2005
|
|
3OKK
 
 | | Crystal structure of S25-39 in complex with Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
2XD9
 
 | | STRUCTURE OF HELICOBACTER PYLORI TYPE II DEHYDROQUINASE IN COMPLEX WITH INHIBITOR COMPOUND (4R,6R,7S)-4,6,7-Trihydroxy-2-((E)-prop-1- enyl)-4,5,6,7-tetrahydrobenzo(b)thiophene-4-carboxylic acid | | Descriptor: | (4R,6R,7S)-4,6,7-TRIHYDROXY-2-[(1E)-PROP-1-EN-1-YL]-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-4-CARBOXYLIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Paz, S, Tizon, L, Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, van Raaij, M.J, Lamb, H, Hawkins, A.R, Lapthorn, A.J, Castedo, L, Gonzalez-Bello, C. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetrahydrobenzothiophene derivatives: conformationally restricted inhibitors of type II dehydroquinase.
ChemMedChem, 6, 2011
|
|
7BGI
 
 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Nelson, N. | | Deposit date: | 2021-01-07 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
2XFQ
 
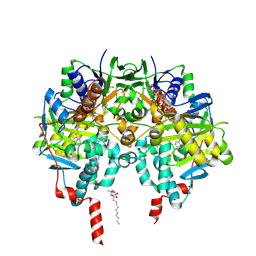 | | Rasagiline-inhibited human monoamine oxidase B in complex with 2-(2- benzofuranyl)-2-imidazoline | | Descriptor: | (1R)-N-(prop-2-en-1-yl)-2,3-dihydro-1H-inden-1-amine, 2-(2-BENZOFURANYL)-2-IMIDAZOLINE, Amine oxidase [flavin-containing] B, ... | | Authors: | Bonivento, D, Milczek, E.M, McDonald, G.R, Binda, C, Holt, A, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potentiation of ligand binding through cooperative effects in monoamine oxidase B.
J. Biol. Chem., 285, 2010
|
|
2ZT9
 
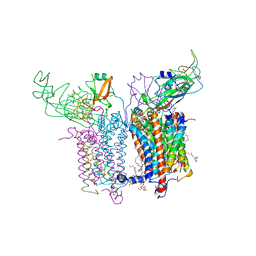 | | Crystal Structure of the Cytochrome b6f Complex from Nostoc sp. PCC 7120 | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, Apocytochrome f, ... | | Authors: | Craner, W.A, Baniulis, D, Yamashita, E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Function, Stability, and Chemical Modification of the Cyanobacterial Cytochrome b6f Complex from Nostoc sp. PCC 7120
J.Biol.Chem., 284, 2009
|
|
2C64
 
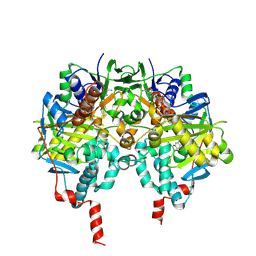 | | MAO inhibition by rasagiline analogues | | Descriptor: | (1R)-6-HYDROXY-N-METHYL-N-[(1Z)-PROP-2-EN-1-YLIDENE]INDAN-1-AMINIUM, AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-11-07 | | Release date: | 2006-01-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of Rasagiline-Related Inhibitors to Human Monoamine Oxidases: A Kinetic and Crystallographic Analysis.
J.Med.Chem., 48, 2005
|
|
4U53
 
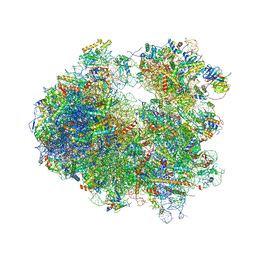 | | Crystal structure of Deoxynivalenol bound to the yeast 80S ribosome | | Descriptor: | (3beta,7alpha)-3,7,15-trihydroxy-12,13-epoxytrichothec-9-en-8-one, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4R
 
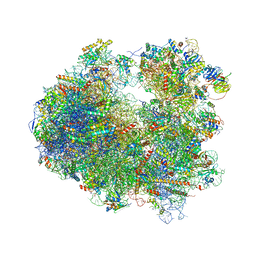 | | Crystal structure of Lactimidomycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R,5S,6E)-2-hydroxy-5-methyl-7-[(2R,3S,4E,6Z,10E)-3-methyl-12-oxooxacyclododeca-4,6,10-trien-2-yl]-4-oxooct-6-en-1-yl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
2ZMH
 
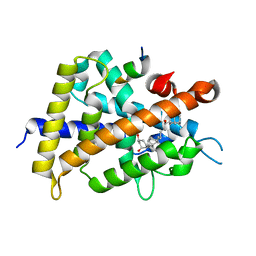 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1R,2E,4R)-4-hydroxy-1-methyl-4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]but-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
2WTX
 
 | | Insight into the mechanism of enzymatic glycosyltransfer with retention through the synthesis and analysis of bisubstrate glycomimetics of trehalose-6-phosphate synthase | | Descriptor: | 1,2-ETHANEDIOL, ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE [UDP-FORMING], ... | | Authors: | Errey, J.C, Lee, S.S, Gibson, R.P, Martinez-Fleites, C, Barry, C.S, Jung, P.M.J, OSullivan, A, Davis, B.G, Davies, G.J. | | Deposit date: | 2009-09-25 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Insight Into Enzymatic Glycosyl Transfer with Retention of Configuration Through Analysis of Glycomimetic Inhibitors.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2Y77
 
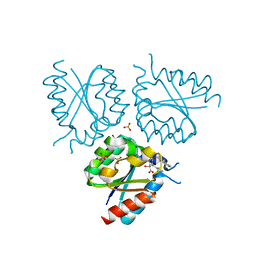 | | Structure of Mycobacterium tuberculosis type II dehydroquinase complexed with (1R,4S,5R)-3-(benzo(b)thiophen-2-ylmethoxy)-1,4,5- trihydroxy-2-(thiophen-2-ylmethyl)cyclohex-2-enecarboxylate | | Descriptor: | (1R,4S,5R)-3-(BENZO[B]THIOPHEN-2-YL)METHOXY-1,4,5-TRIHYDROXY-2-(THIEN-2-YL)METHYLCYCLOHEX-2-EN-1-CARBOXYLATE, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Fox, G.C, Tizon, L, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Ainsa, J.A, Castedo, L, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A prodrug approach for improving antituberculosis activity of potent Mycobacterium tuberculosis type II dehydroquinase inhibitors.
J. Med. Chem., 54, 2011
|
|
2YG2
 
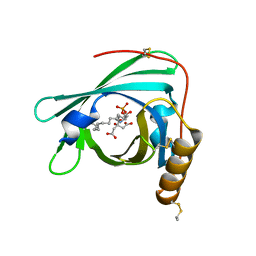 | | Structure of apolioprotein M in complex with Sphingosine 1-Phosphate | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, APOLIPOPROTEIN M, CITRATE ANION | | Authors: | Christoffersen, C, Obinata, H, Gowda, S, Galvani, S, Ahnstrom, J, Sevvana, M, Egerer-Sieber, C, Muller, Y.A, Hla, T, Nielsen, L, Dahlback, B. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Endothelium-Protective Sphingosine-1-Phosphate Provided by Hdl-Associated Apolipoprotein M.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2CHM
 
 | | Crystal structure of N2 substituted pyrazolo pyrimidinones - a flipped binding mode in PDE5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2-(BUT-3-EN-1-YLOXY)-5-(1-HYDROXYVINYL)PYRIDIN-3-YL]-3-ETHYL-2-(1-ETHYLAZETIDIN-3-YL)-1,2,6,7A-TETRAHYDRO-7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, CGMP-SPECIFIC 3', ... | | Authors: | Allerton, C.M.N, Barber, C.G, Beaumont, K.C, Brown, D.G, Cole, S.M, Ellis, D, Lane, C.A.L, Maw, G.N, Mount, N.M, Rawson, D.J, Robinson, C.M, Street, S.D.A, Summerhill, N.W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-06-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Novel Series of Potent and Selective Pde5 Inhibitors with Potential for High and Dose-Independent Oral Bioavailability
J.Med.Chem., 49, 2006
|
|
2ZMJ
 
 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1S,2E,5R)-5-hydroxy-1-methyl-6-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hex-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
2ZK5
 
 | | Human peroxisome proliferator-activated receptor gamma ligand binding domain complexed with nitro-233 | | Descriptor: | 3-[5-(2-nitropent-1-en-1-yl)furan-2-yl]benzoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Shiraki, T, Oyama, T, Fujimoto, Y, Morikawa, K. | | Deposit date: | 2008-03-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids
J.Mol.Biol., 385, 2009
|
|
3OKN
 
 | | Crystal structure of S25-39 in complex with Kdo(2.4)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
2ZYP
 
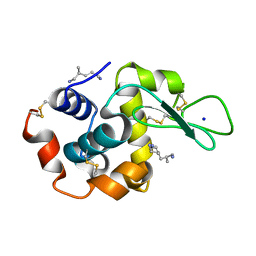 | | X-ray structure of hen egg-white lysozyme with poly(allyl amine) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Ito, L, Tomita, S, Yamaguchi, H, Shiraki, K. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | X-ray structure of hen egg-white lysozyme with poly(allyl amine)
to be published
|
|
2AOB
 
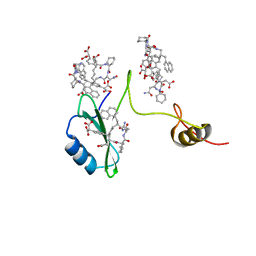 | | Crystal structures of a high-affinity macrocyclic peptide mimetic in complex with the Grb2 SH2 domain | | Descriptor: | 2-(4-((9S,10S,14S,Z)-18-(2-AMINO-2-OXOETHYL)-9-(CARBOXYMETHYL)-14-(NAPHTHALEN-1-YLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL)PHENYL)MALONIC ACID, Growth factor receptor-bound protein 2 | | Authors: | Phan, J, Shi, Z.D, Burke, T.R, Waugh, D.S. | | Deposit date: | 2005-08-12 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a High-affinity Macrocyclic Peptide Mimetic in Complex with the Grb2 SH2 Domain.
J.Mol.Biol., 353, 2005
|
|
2ZXC
 
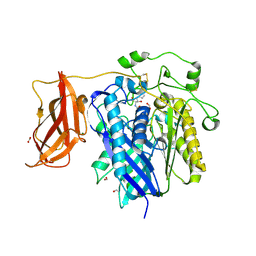 | | Ceramidase complexed with C2 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Okano, H, Inoue, T, Okino, N, Kakuta, Y, Matsumura, H, Ito, M. | | Deposit date: | 2008-12-22 | | Release date: | 2009-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
7LI5
 
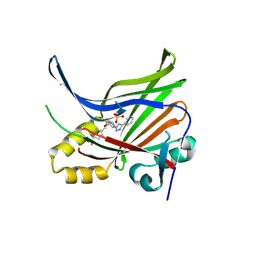 | | Crystal Structure Analysis of human TEAD1 | | Descriptor: | 1-[(3R,4R)-3-[4-(pyridin-3-yl)-1H-1,2,3-triazol-1-yl]-4-{[4-(trifluoromethyl)phenyl]methoxy}pyrrolidin-1-yl]prop-2-en-1-one, SULFATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure Analysis of human TEAD1
To Be Published
|
|
1QLU
 
 | |
1Q20
 
 | | Crystal Structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of PAP and pregnenolone | | Descriptor: | (3BETA)-3-HYDROXYPREGN-5-EN-20-ONE, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Lee, K.A, Fuda, H, Lee, Y.C, Negishi, M, Strott, C.A, Pedersen, L.C. | | Deposit date: | 2003-07-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone
and 3'-phosphoadenosine 5'-phosphate. Rationale for specificity differences between prototypical
SULT2A1 and the SULT2BG1 isoforms.
J.Biol.Chem., 278, 2003
|
|
1S2Q
 
 | | Crystal structure of MAOB in complex with N-propargyl-1(R)-aminoindan (Rasagiline) | | Descriptor: | (1R)-N-(prop-2-en-1-yl)-2,3-dihydro-1H-inden-1-amine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-30 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structures of Monoamine Oxidase B in Complex with Four Inhibitors of the N-Propargylaminoindan Class.
J.Med.Chem., 47, 2004
|
|
1R1K
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Billas, I.M.L, Iwema, T, Garnier, J.-M, Mitschler, A, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-24 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Nature, 426, 2003
|
|
