6RGY
 
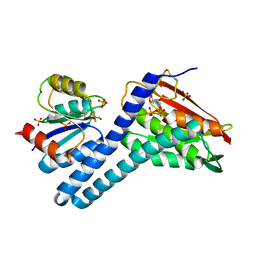 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
8G3F
 
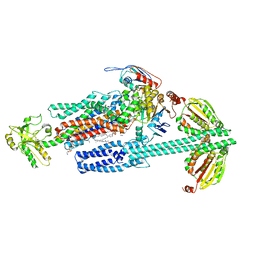 | | BceAB-S nucleotide free BceS state 1 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G3B
 
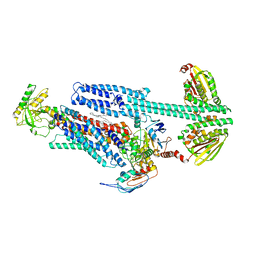 | | BceAB-S nucleotide free TM state 2 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
6RH2
 
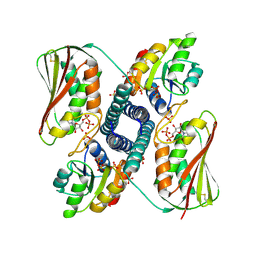 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RKS
 
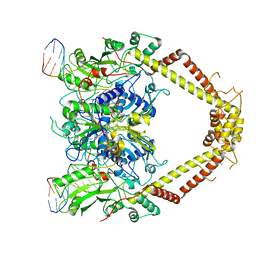 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 without TOPRIM insertion | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
8G3L
 
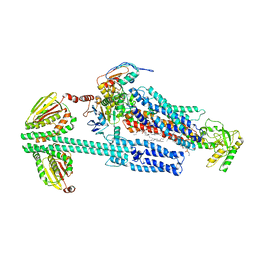 | | BceAB-S nucleotide free BceS state 2 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-08 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G4C
 
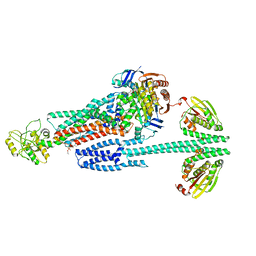 | | BceABS ATPgS high res TM | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G4D
 
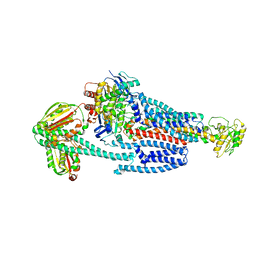 | | BceABS ATPgS tilted BceS | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
6RH7
 
 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
8G3A
 
 | | BceAB-S nucleotide free TM state 1 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
6RH0
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 5.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RKV
 
 | |
6RKU
 
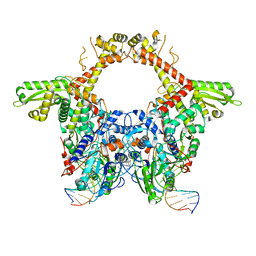 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RFV
 
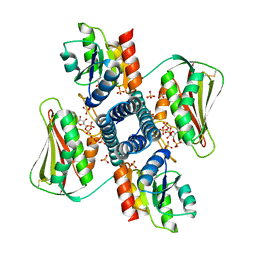 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RKW
 
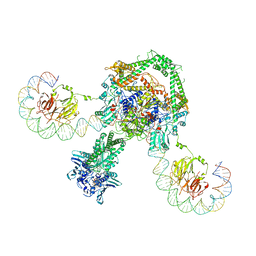 | | CryoEM structure of the complete E. coli DNA Gyrase complex bound to a 130 bp DNA duplex | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (58-MER), DNA (62-MER), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6S1K
 
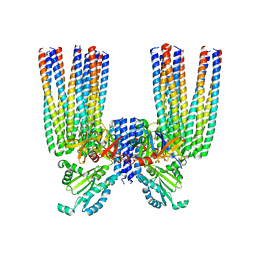 | | E. coli Core Signaling Unit, carrying QQQQ receptor mutation | | Descriptor: | CheW, Chemotaxis protein CheA, Methyl-accepting chemotaxis protein I | | Authors: | Cassidy, C.K. | | Deposit date: | 2019-06-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.38 Å) | | Cite: | Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations.
Commun Biol, 3, 2020
|
|
6RH1
 
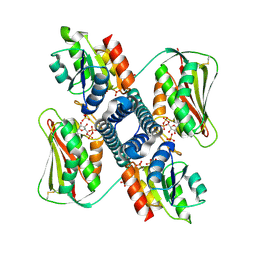 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 D53A pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
4HY1
 
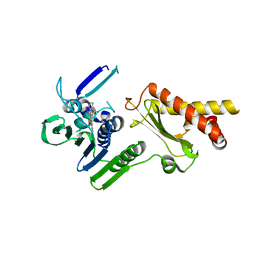 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 6-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)-1H-isoindol-1-one, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I3H
 
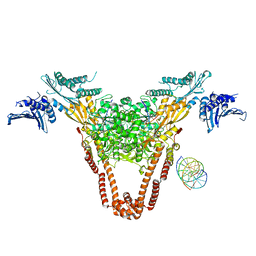 | | A three-gate structure of topoisomerase IV from Streptococcus pneumoniae | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*GP*CP*GP*GP*TP*AP*AP*TP*AP*CP*GP*GP*TP*TP*AP*TP*CP*CP*AP*CP*AP*GP*AP*AP*TP*CP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*AP*TP*TP*CP*TP*GP*TP*GP*GP*AP*TP*AP*AP*CP*CP*GP*TP*AP*TP*TP*AP*CP*CP*GP*CP*CP*TP*TP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Crevel, I, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2012-11-26 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of an 'open' clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport.
Nucleic Acids Res., 41, 2013
|
|
4HXZ
 
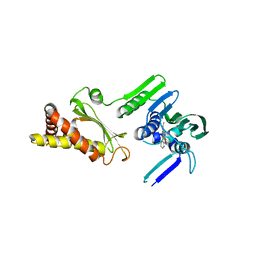 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 6-ethyl-4-methoxy-2-(pyridin-3-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbaldehyde, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HYM
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 7-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)pyrido[2,3-b]pyrazin-2(1H)-one, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JAU
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JPB
 
 | | The structure of a ternary complex between CheA domains P4 and P5 with CheW and with an unzipped fragment of TM14, a chemoreceptor analog from Thermotoga maritima. | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein | | Authors: | Li, X, Bayas, C, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2013-03-19 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | The 3.2 angstrom resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays.
Biochemistry, 52, 2013
|
|
4JAS
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E and RR468mutant V13P, L14I, I17M and N21V) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase, MAGNESIUM ION, ... | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JUO
 
 | | A low-resolution three-gate structure of topoisomerase IV from Streptococcus pneumoniae in space group H32 | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA topoisomerase 4 subunit A, DNA topoisomerase 4 subunit B, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Crevel, I, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.53 Å) | | Cite: | Structure of an 'open' clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport.
Nucleic Acids Res., 41, 2013
|
|
