6SFQ
 
 | | Atomic resolution structure of human Carbonic Anhydrase II in complex with (R)-5-phenyloxazolidine-2,4-dione | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, (5R)-5-phenyl-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of human Carbonic Anhydrase II in complex with (R)-5-phenyloxazolidine-2,4-dione
To Be Published
|
|
8GT7
 
 | | Structure of falcipain and human Stefin A mutant complex | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Cystatin-A, ... | | Authors: | Chakraborty, S, Biswas, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-09-13 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structure of falcipain and human Stefin A complex
To Be Published
|
|
7KSS
 
 | | DNA Polymerase Mu, dGTP:Ct Pre-Catalytic Ground State Ternary Complex, 10 mM Ca2+ (20min) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
2R9W
 
 | | AmpC beta-lactamase with bound Phthalamide inhibitor | | Descriptor: | 2-[(1R)-1-carboxy-2-naphthalen-1-ylethyl]-1,3-dioxo-2,3-dihydro-1H-isoindole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Babaoglu, K, Shoichet, B.K. | | Deposit date: | 2007-09-13 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase.
J.Med.Chem., 51, 2008
|
|
3KN2
 
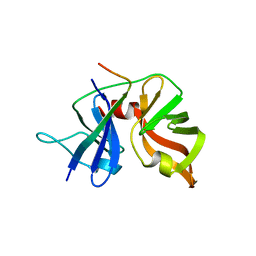 | | HCV NS3 Protease Domain with ketoamide inhibitor | | Descriptor: | (1R,2S,5S)-3-{(2S)-2-(2,3-dihydro-1H-inden-2-yl)-2-[({(1S)-2,2-dimethyl-1-[(2-oxopiperidin-1-yl)methyl]propyl}carbamoyl)amino]acetyl}-6,6-dimethyl-N-{(1S)-1-[oxo(prop-2-en-1-ylamino)acetyl]butyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, HCV NS3 Protease Domain, Peptide KK-NS4A-KK, ... | | Authors: | Nair, L.G, Sannigrahi, M, Pinto, P, Bogen, S, Chen, K.X, Njoroge, G, Prongay, A. | | Deposit date: | 2009-11-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | P4 capped amides and lactams as HCV NS3 protease inhibitors with improved potency and DMPK profile.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7KTJ
 
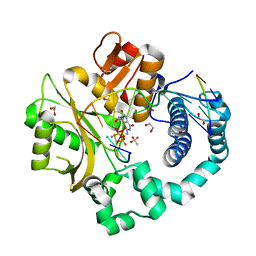 | | DNA Polymerase Mu (K438D), 8-oxodGTP:Ct Pre-Catalytic Ground State Ternary Complex, 20 mM Ca2+ (120min) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
4MBY
 
 | | Structure of B-Lymphotropic Polyomavirus VP1 in complex with 3'-sialyllactose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Khan, Z.M, Neu, U, Stehle, T. | | Deposit date: | 2013-08-21 | | Release date: | 2013-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structures of B-Lymphotropic Polyomavirus VP1 in Complex with Oligosaccharide Ligands.
Plos Pathog., 9, 2013
|
|
7KVC
 
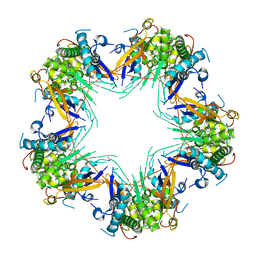 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
2RBD
 
 | |
7KVD
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
8XX0
 
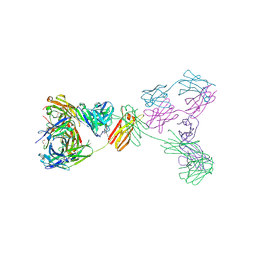 | | Crystal structure of anti-IgE antibody HMK-12 Fab complexed with IgE F(ab')2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SPE7 immunoglobulin E F(ab')2 heavy chain, ... | | Authors: | Hirano, T, Koyanagi, A, Kasai, M, Okumura, K. | | Deposit date: | 2024-01-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric inhibition of IgE-Fc epsilon RI interactions by simultaneous targeting of IgE F(ab')2 epitopes.
Commun Biol, 7, 2024
|
|
2RC7
 
 | |
8YBK
 
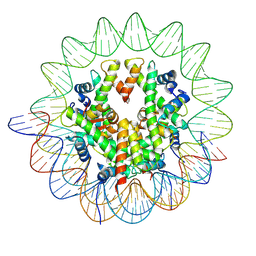 | | Cryo-EM structure of the human nucleosome containing the H3.1 E97K mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
9H6P
 
 | | Crystal structure of NUDT14 complexed with novel compound AMNUDT14-003 in spacegroup P1 | | Descriptor: | 3-(1H-indol-6-yl)-1-(1-methylpiperidin-4-yl)pyrazolo[3,4-d]pyrimidin-4-amine, Uridine diphosphate glucose pyrophosphatase NUDT14 | | Authors: | Balikci, E, Koekemoer, L, Dlamini, L.S, Gurav, N, Apostolidou, M, Adcock, C, McGown, A, Marques, A.M.C, Spencer, J, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-10-24 | | Release date: | 2025-11-05 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Crystal structure of NUDT14 complexed with novel compound AMNUDT14-003 in spacegroup
To Be Published
|
|
6SI3
 
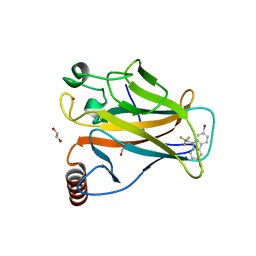 | | p53 cancer mutant Y220S in complex with small-molecule stabilizer PK9301 | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-bromanyl-9-[2,2,2-tris(fluoranyl)ethyl]carbazol-3-yl]-~{N}-methyl-methanamine, Cellular tumor antigen p53, ... | | Authors: | Joerger, A.C, Bauer, M.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
8Y37
 
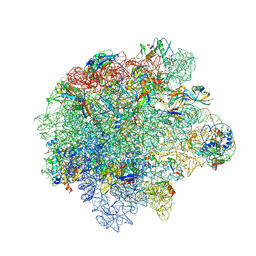 | | Cryo-EM structure of Staphylococcus aureus (15B196) 50S ribosome in complex with MCX-190. | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, 7-[4-[3-[[(1~{S},2~{R},5~{R},6~{S},7~{S},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-9-methoxy-1,5,7,9,11,13-hexamethyl-4,12,16-tris(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-6-yl]oxycarbonylamino]propoxy]but-1-ynyl]-1-methyl-4-oxidanylidene-quinoline-3-carboxylic acid, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
8Y36
 
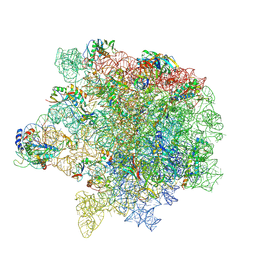 | | cryo-EM structure of Staphylococcus aureus(ATCC 29213) 50S ribosome in complex with MCX-190. | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, 7-[4-[3-[[(1~{S},2~{R},5~{R},6~{S},7~{S},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-9-methoxy-1,5,7,9,11,13-hexamethyl-4,12,16-tris(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-6-yl]oxycarbonylamino]propoxy]but-1-ynyl]-1-methyl-4-oxidanylidene-quinoline-3-carboxylic acid, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
3T8S
 
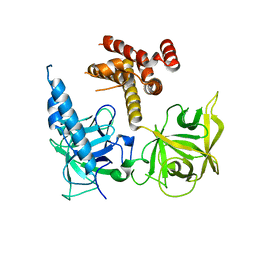 | | Apo and InsP3-bound Crystal Structures of the Ligand-Binding Domain of an InsP3 Receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Lin, C, Baek, K, Lu, Z. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Apo and InsP(3)-bound crystal structures of the ligand-binding domain of an InsP(3) receptor.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8G66
 
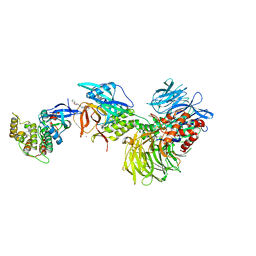 | | Structure with SJ3149 | | Descriptor: | (3S)-3-{5-[(1,2-benzoxazol-3-yl)amino]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}piperidine-2,6-dione, Casein kinase I isoform alpha, DNA damage-binding protein 1, ... | | Authors: | Miller, D.J, Young, S.M, Fischer, M. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of ternary complex with molecular glue targeting CK1A for degradation by the CRL4CRBN ubiquitin ligase
To Be Published
|
|
5LQQ
 
 | | Structure of Autotaxin (ENPP2) with LM350 | | Descriptor: | 3-(6-chloranyl-2-methyl-1-phenyl-indol-3-yl)sulfanylbenzoic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.J, Heidebrecht, T, Castelmur, E, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-08-17 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Activity Relationships of Small Molecule Autotaxin Inhibitors with a Discrete Binding Mode.
J. Med. Chem., 60, 2017
|
|
8XRP
 
 | | The Cryo-EM structure of IL-12, receptor subunit beta-1 and receptor subunit beta-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, Interleukin-12 receptor subunit beta-2, ... | | Authors: | Chen, H.Q, Wang, X.Q, Ge, X.F, Zeng, J.W, Wang, J.W. | | Deposit date: | 2024-01-07 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure and assembly of the human IL-12 signaling complex.
Structure, 32, 2024
|
|
5CD3
 
 | | Structure of immature VRC01-class antibody DRVIA7 | | Descriptor: | DRVIA7 Heavy Chain, DRVIA7 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor.
Immunity, 44, 2016
|
|
6SPP
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 | | Descriptor: | CITRIC ACID, GLYCEROL, Methionine--tRNA ligase, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
7LG4
 
 | |
6SR1
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 35 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
