5VZ9
 
 | | Post-catalytic complex of human Polymerase Mu (G433A) mutant with incoming dTTP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5W1C
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5VZG
 
 | | Pre-catalytic ternary complex of human Polymerase Mu (W434H) mutant with incoming nonhydrolyzable UMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, CHLORIDE ION, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5W0W
 
 | | Crystal structure of Protein Phosphatase 2A bound to TIPRL | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform, ... | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
5W1Z
 
 | | Crystal Structure of inosine-substituted decamer duplex DNA (I4) | | Descriptor: | DNA (5'-D(*CP*CP*AP*IP*IP*CP*CP*(BRU)P*IP*I)-3'), MAGNESIUM ION, SODIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparative analysis of inosine-substituted duplex DNA by circular dichroism and X-ray crystallography.
J. Biomol. Struct. Dyn., 36, 2018
|
|
5W2H
 
 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase in complex with Ins(1,4,5)P3 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2017-06-06 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural features of human inositol phosphate multikinase rationalize its inositol phosphate kinase and phosphoinositide 3-kinase activities.
J. Biol. Chem., 292, 2017
|
|
5W36
 
 | |
5LRA
 
 | | Plastidial phosphorylase PhoI from barley in complex with maltotetraose | | Descriptor: | Alpha-1,4 glucan phosphorylase, PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Krucewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
5W3L
 
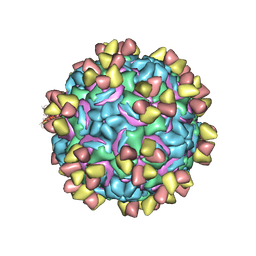 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (4 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W40
 
 | |
5LS2
 
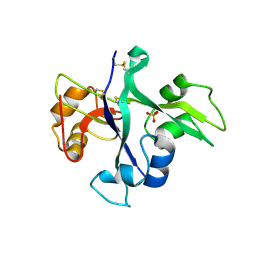 | | Receptor mediated chitin perception in legumes is functionally seperable from Nod factor perception | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase, SULFATE ION | | Authors: | Bozsoki, Z, Cheng, J, Feng, F, Gysel, K, Andersen, K.R, Oldroyd, G, Blaise, M, Radutoiu, S, Stougaard, J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W6F
 
 | |
5W6V
 
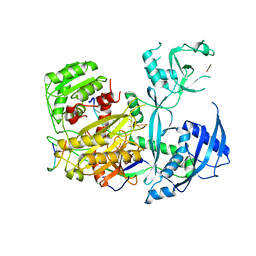 | |
5LNW
 
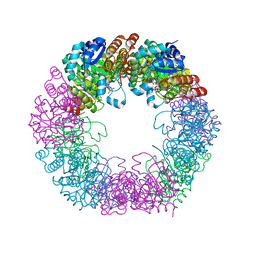 | | Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, GLYCEROL, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5W0V
 
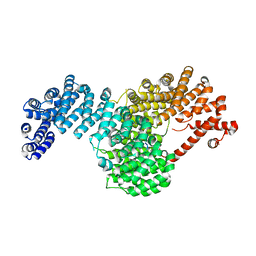 | | Crystal structure of full-length Kluyveromyces lactis Kap123 with histone H4 1-34 | | Descriptor: | Histone H4 1-34, Kap123 | | Authors: | An, S, Yoon, J, Song, J.-J, Cho, U.-S. | | Deposit date: | 2017-05-31 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123.
Elife, 6, 2017
|
|
5W1T
 
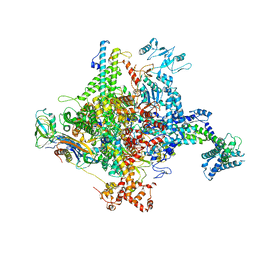 | |
5W5Z
 
 | | Crystal structure of BAXP168G in complex with an activating antibody at high resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C10 Fab heavy chain, ... | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
5W1W
 
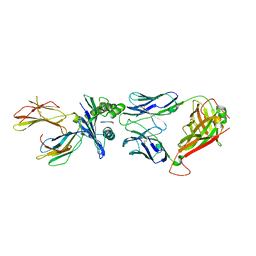 | | Structure of the HLA-E-VMAPRTLVL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
5W4E
 
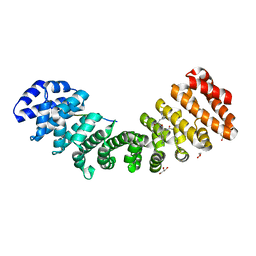 | | Importin binding to Tdt NLS peptide | | Descriptor: | GLYCEROL, Importin subunit alpha-1,Importin subunit alpha-1, human DNA repair polymerase Tdt | | Authors: | Pedersen, L.C, London, R. | | Deposit date: | 2017-06-10 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Variations in nuclear localization strategies among pol X family enzymes.
Traffic, 2018
|
|
7R6A
 
 | |
5W3E
 
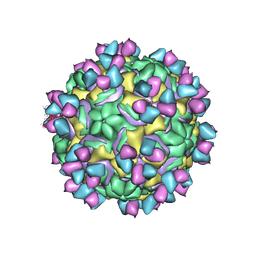 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3Y
 
 | |
5W4H
 
 | |
5W4I
 
 | |
5M0D
 
 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2,Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, GLYCEROL, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
