1DDO
 
 | |
1DDP
 
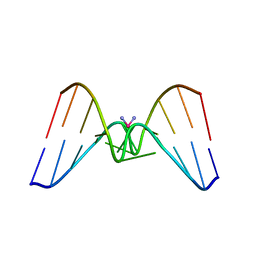 | | Solution structure of a CISPLATIN-INDUCED [CATAGCTATG]2 Interstrand cross-link | | Descriptor: | Cisplatin, DNA (5'-D(*CP*AP*TP*AP*GP*CP*TP*AP*TP*G)-3') | | Authors: | Zhu, L, Huang, H, Reid, B.R, Drobny, G.P, Hopkins, P.B. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cisplatin-induced DNA interstrand cross-link.
Science, 270, 1995
|
|
1DDR
 
 | |
1DDS
 
 | |
1DDT
 
 | |
1DDU
 
 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH CB3717 AND 2',5'-DIDEOXYURIDINE (DDURD) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-5'DIDEOXYURIDINE, PHOSPHATE ION, ... | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1DDV
 
 | | CRYSTAL STRUCTURE OF THE HOMER EVH1 DOMAIN WITH BOUND MGLUR PEPTIDE | | Descriptor: | GLGF-DOMAIN PROTEIN HOMER, METABOTROPIC GLUTAMATE RECEPTOR MGLUR5 | | Authors: | Beneken, J, Tu, J.C, Xiao, B, Worley, P.F, Leahy, D.J. | | Deposit date: | 1999-11-11 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition.
Neuron, 26, 2000
|
|
1DDW
 
 | | HOMER EVH1 DOMAIN UNLIGANDED | | Descriptor: | GLGF-DOMAIN PROTEIN HOMER | | Authors: | Beneken, J, Tu, J.C, Xiao, B, Worley, P.F, Leahy, D.J. | | Deposit date: | 1999-11-11 | | Release date: | 2000-05-10 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition.
Neuron, 26, 2000
|
|
1DDX
 
 | | CRYSTAL STRUCTURE OF A MIXTURE OF ARACHIDONIC ACID AND PROSTAGLANDIN BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2: PROSTAGLANDIN STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[6-(3-HYDROPEROXY-OCT-1-ENYL)-2,3-DIOXA-BICYCLO[2.2.1]HEPT-5-YL]-HEPT-5-ENOIC ACID, ... | | Authors: | Kiefer, J.R, Pawlitz, J.L, Moreland, K.T, Stegeman, R.A, Gierse, J.K, Stevens, A.M, Goodwin, D.C, Rowlinson, S.W, Marnett, L.J, Stallings, W.C, Kurumbail, R.G. | | Deposit date: | 1999-11-11 | | Release date: | 2000-05-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the stereochemistry of the cyclooxygenase reaction.
Nature, 405, 2000
|
|
1DDY
 
 | |
1DDZ
 
 | | X-RAY STRUCTURE OF A BETA-CARBONIC ANHYDRASE FROM THE RED ALGA, PORPHYRIDIUM PURPUREUM R-1 | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Mitsuhashi, S, Mizushima, T, Yamashita, E, Miyachi, S, Tsukihara, T. | | Deposit date: | 1999-11-12 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of beta-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO(2) hydration.
J.Biol.Chem., 275, 2000
|
|
1DE0
 
 | |
1DE1
 
 | |
1DE2
 
 | |
1DE3
 
 | | SOLUTION STRUCTURE OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | RIBONUCLEASE ALPHA-SARCIN | | Authors: | Perez-Canadillas, J.M, Campos-Olivas, R, Santoro, J, Lacadena, J, Martinez del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 1999-11-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity.
J.Mol.Biol., 299, 2000
|
|
1DE4
 
 | | HEMOCHROMATOSIS PROTEIN HFE COMPLEXED WITH TRANSFERRIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-2-MICROGLOBULIN, CALCIUM ION, ... | | Authors: | Bennett, M.J, Lebron, J.A, Bjorkman, P.J. | | Deposit date: | 1999-11-12 | | Release date: | 2000-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor.
Nature, 403, 2000
|
|
1DE5
 
 | | L-RHAMNOSE ISOMERASE | | Descriptor: | L-RHAMNITOL, L-RHAMNOSE ISOMERASE, ZINC ION | | Authors: | Korndorfer, I.P, Fessner, W.D, Matthews, B.W. | | Deposit date: | 1999-11-12 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution.
J.Mol.Biol., 300, 2000
|
|
1DE6
 
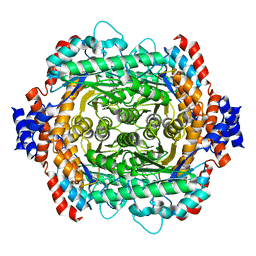 | | L-RHAMNOSE ISOMERASE | | Descriptor: | L-RHAMNOSE, L-RHAMNOSE ISOMERASE, MANGANESE (II) ION, ... | | Authors: | Korndorfer, I.P, Fessner, W.D, Matthews, B.W. | | Deposit date: | 1999-11-13 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution.
J.Mol.Biol., 300, 2000
|
|
1DE7
 
 | |
1DE8
 
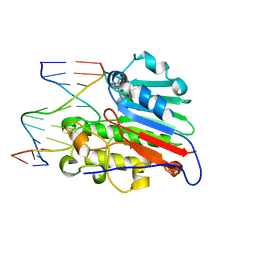 | | HUMAN APURINIC/APYRIMIDINIC ENDONUCLEASE-1 (APE1) BOUND TO ABASIC DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*CP*(3DR)P*GP*AP*TP*CP*G)-3'), MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected
Nature, 403, 2000
|
|
1DE9
 
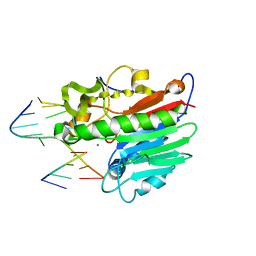 | | HUMAN APE1 ENDONUCLEASE WITH BOUND ABASIC DNA AND MN2+ ION | | Descriptor: | 5'-d(*CP*TP*AP*C)-3', 5'-d(*GP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-d(P*(3DR)P*GP*AP*TP*C)-3', ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
1DEA
 
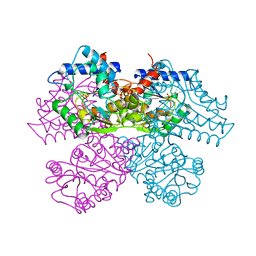 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1DEB
 
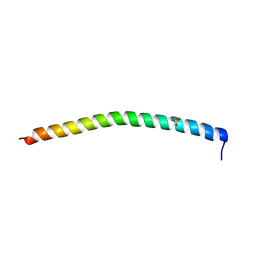 | |
1DEC
 
 | |
1DED
 
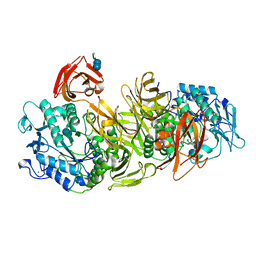 | | CRYSTAL STRUCTURE OF ALKALOPHILIC ASPARAGINE 233-REPLACED CYCLODEXTRIN GLUCANOTRANSFERASE COMPLEXED WITH AN INHIBITOR, ACARBOSE, AT 2.0 A RESOLUTION | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Ishii, N, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 1999-11-14 | | Release date: | 2000-04-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alkalophilic asparagine 233-replaced cyclodextrin glucanotransferase complexed with an inhibitor, acarbose, at 2.0 A resolution.
J.Biochem.(Tokyo), 127, 2000
|
|
