4G3Y
 
 | | Crystal structure of TNF-alpha in complex with Infliximab Fab fragment | | Descriptor: | Tumor necrosis factor, infliximab Fab H, infliximab Fab L | | Authors: | Liang, S.Y, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-07-15 | | Release date: | 2013-03-27 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for treating tumor necrosis factor alpha (TNFalpha)-associated diseases with the therapeutic antibody infliximab
J.Biol.Chem., 288, 2013
|
|
3LDL
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
4BNG
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-pentyl- 2-phenoxyphenol | | Descriptor: | 5-PENTYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
4PCH
 
 | |
4BZ6
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with SAHA | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, HISTONE DEACETYLASE 8, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4C2Y
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA co-factor | | Descriptor: | CITRIC ACID, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE 1, ... | | Authors: | Thinon, E, Serwa, R.A, Brannigan, J.A, Brassat, U, Wright, M.H, Heal, W.P, Wilkinson, A.J, Mann, D.J, Tate, E.W. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Global Profiling of Co- and Post-Translationally N-Myristoylated Proteomes in Human Cells.
Nat.Commun., 5, 2014
|
|
3LRB
 
 | | Structure of E. coli AdiC | | Descriptor: | Arginine/agmatine antiporter | | Authors: | Gao, X, Lu, F, Zhou, L, Shi, Y. | | Deposit date: | 2010-02-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Structure and mechanism of an amino acid antiporter
Science, 324, 2009
|
|
4BWP
 
 | | Structure of Drosophila Melanogaster PAN3 pseudokinase | | Descriptor: | AMP PHOSPHORAMIDATE, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN-3 | | Authors: | Christie, M, Boland, A, Huntzinger, E, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2013-07-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the Pan3 Pseudokinase Reveals the Basis for Interactions with the Pan2 Deadenylase and the Gw182 Proteins
Mol.Cell, 51, 2013
|
|
4A1G
 
 | | The crystal structure of the human Bub1 TPR domain in complex with the KI motif of Knl1 | | Descriptor: | MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1, PROTEIN CASC5 | | Authors: | Krenn, V, Wehenkel, A, Li, X, Santaguida, S, Musacchio, A. | | Deposit date: | 2011-09-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis Reveals Features of the Spindle Checkpoint Kinase Bub1-Kinetochore Subunit Knl1 Interaction.
J.Cell Biol., 196, 2012
|
|
4A1V
 
 | | Co-Complex structure of NS3-4A protease with the optimized inhibitory peptide CP5-46A-4D5E | | Descriptor: | CHLORIDE ION, CP5-46A-4D5E, NON-STRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
3LKH
 
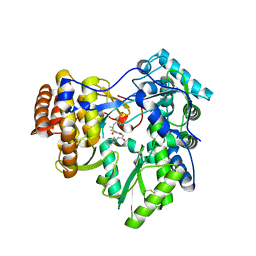 | |
4DIA
 
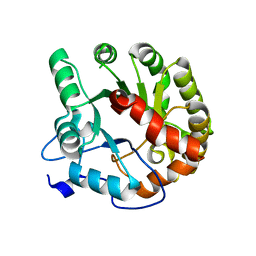 | | CRYSTAL STRUCTURE OF THE D248N mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 4.6 | | Descriptor: | 2-pyrone-4,6-dicarbaxylate hydrolase | | Authors: | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4DT6
 
 | | Co-crystal structure of eIF4E with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-(4-chlorophenoxy)ethyl]guanosine 5'-(dihydrogen phosphate), Eukaryotic translation initiation factor 4E, ... | | Authors: | Min, X, Johnstone, S, Walker, N, Wang, Z. | | Deposit date: | 2012-02-20 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Design, Synthesis, and Evaluation of Guanine-Derived Inhibitors of the eIF4E mRNA-Cap Interaction.
J.Med.Chem., 55, 2012
|
|
4ALI
 
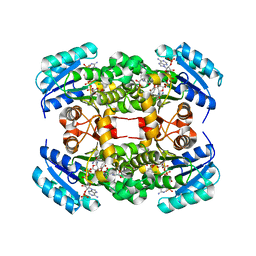 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P1) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
3LBY
 
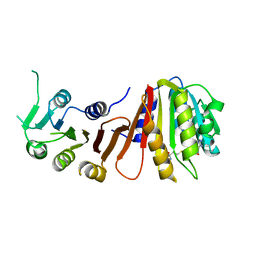 | |
4DGR
 
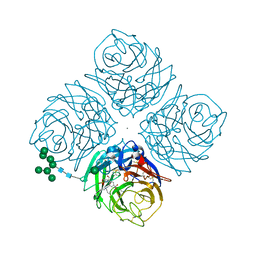 | | Influenza Subtype 9 Neuraminidase Benzoic Acid Inhibitor Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2,2-bis(hydroxymethyl)-5-oxopyrrolidin-1-yl]-3-[(dipropylamino)methyl]benzoic acid, CALCIUM ION, ... | | Authors: | Venkatramani, L, Johnson, E, Kolavi, G, Air, G.M, Brouillette, W, Mooers, B.H.M. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a new benzoic acid inhibitor of influenza neuraminidase bound with a new tilt induced by overpacking sub-site C6.
Bmc Struct.Biol., 12, 2012
|
|
4AL3
 
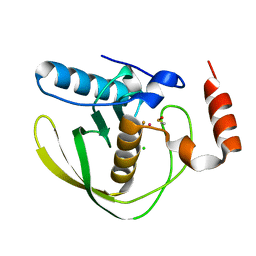 | |
4DHM
 
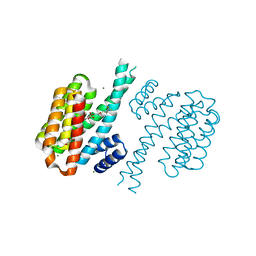 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHT
 
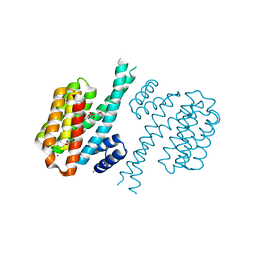 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 PROTEIN SIGMA, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4AOZ
 
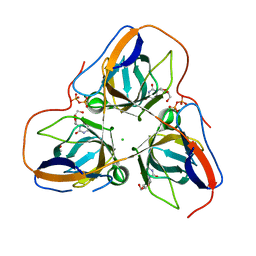 | | B. subtilis dUTPase YncF in complex with dU, PPi and Mg (P212121) | | Descriptor: | 2'-DEOXYURIDINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Timm, J, Garcia-Nafria, J, Harrison, C, Turkenburg, J.P, Wilson, K.S. | | Deposit date: | 2012-03-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tying Down the Arm in Bacillus Dutpase: Structure and Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3LSD
 
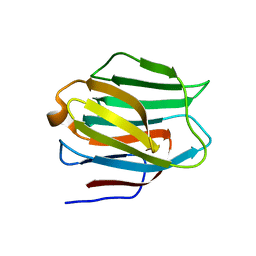 | | N-Domain of human adhesion/growth-regulatory galectin-9 | | Descriptor: | Galectin-9 | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2010-02-12 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | N-domain of human adhesion/growth-regulatory galectin-9: preference for distinct conformers and non-sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution.
Int.J.Biochem.Cell Biol., 42, 2010
|
|
4A3S
 
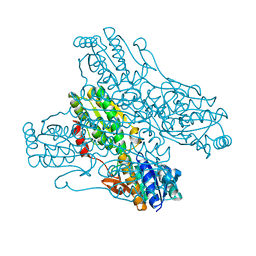 | | Crystal structure of PFK from Bacillus subtilis | | Descriptor: | 6-PHOSPHOFRUCTOKINASE | | Authors: | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A.S, Harwood, C.R, Lewis, R.J. | | Deposit date: | 2011-10-04 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissection of the Network of Interactions that Links RNA Processing with Glycolysis in the Bacillus Subtilis Degradosome.
J.Mol.Biol., 416, 2012
|
|
4P1G
 
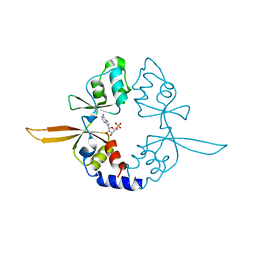 | | Crystal structure of the Bateman domain of murine magnesium transporter CNNM2 bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Abascal-Palacios, G, Diercks, T, Oyenarte, I, Ereno-Orbea, J, Encinar, J.A, Spiwok, V, Terashima, H, Accardi, A, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To Be Published
|
|
4P4R
 
 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between Allergic Hypersensitivity and Autoimmunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DP alpha 1 chain, ... | | Authors: | Clayton, G.M, Crawford, F, Kappler, J.W. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
3LU2
 
 | | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase | | Descriptor: | Lmo2462 protein, ZINC ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase
To be Published
|
|
