1M6Z
 
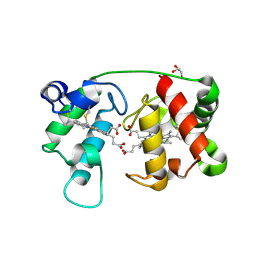 | | Crystal structure of reduced recombinant cytochrome c4 from Pseudomonas stutzeri | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome c4, GLYCEROL, ... | | Authors: | Noergaard, A, Harris, P, Larsen, S, Christensen, H.E.M. | | Deposit date: | 2002-07-18 | | Release date: | 2003-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural comparison of recombinant Pseudomonas stutzeri cytochrome c4 in two oxidation states
To be Published
|
|
8PD6
 
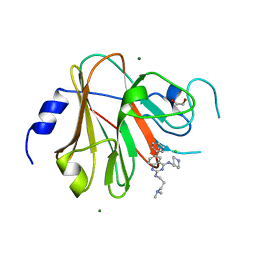 | | Crystal structure of the TRIM58 PRY-SPRY domain in complex with TRIM-473 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase TRIM58, ... | | Authors: | Renatus, M, Hoegenauer, K, Schroeder, M. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Ligands for TRIM58, a Novel Tissue-Selective E3 Ligase.
Acs Med.Chem.Lett., 14, 2023
|
|
6SSA
 
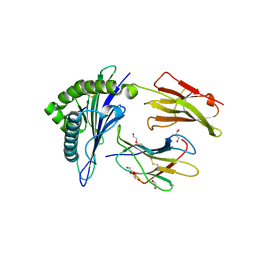 | | Human Leukocyte Antigen Class I A02 Carrying LLWNGPMQV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CALCIUM ION, ... | | Authors: | Rizkallah, P.J, Bovay, A. | | Deposit date: | 2019-09-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of a superagonist variant of the immunodominant Yellow fever virus epitope NS4b214-222by combinatorial peptide library screening.
Mol.Immunol., 125, 2020
|
|
8I48
 
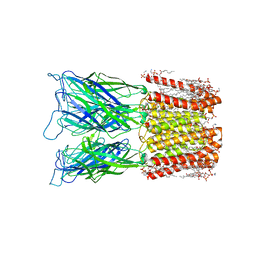 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-01-18 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
4FT4
 
 | |
6SWM
 
 | | selenol bound carbonic anhydrase I | | Descriptor: | (2~{R})-1-prop-2-enoxy-3-selanyl-propan-2-ol, Carbonic anhydrase 1, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-07 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Selenols: a new class of carbonic anhydrase inhibitors.
Chem.Commun.(Camb.), 55, 2019
|
|
2OUO
 
 | | Crystal Structure of the Bromo domain 2 in human Bromodomain Containing Protein 4 (BRD4) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Keates, T, Savitsky, P, Burgess, N, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
6SS9
 
 | | Human Leukocyte Antigen Class I A02 Carrying LLWNGPMHV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rizkallah, P.J, Bovay, A. | | Deposit date: | 2019-09-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a superagonist variant of the immunodominant Yellow fever virus epitope NS4b214-222by combinatorial peptide library screening.
Mol.Immunol., 125, 2020
|
|
3MU7
 
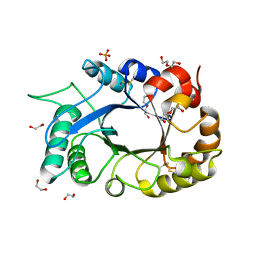 | | Crystal structure of the xylanase and alpha-amylase inhibitor protein (XAIP-II) from scadoxus multiflorus at 1.2 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, xylanase and alpha-amylase inhibitor protein | | Authors: | Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-02 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Modulation of inhibitory activity of xylanase-alpha-amylase inhibitor protein (XAIP): binding studies and crystal structure determination of XAIP-II from Scadoxus multiflorus at 1.2 A resolution.
Bmc Struct.Biol., 10, 2010
|
|
3VFE
 
 | | Virtual Screening and X-Ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group | | Descriptor: | 4-{[(3R)-3-{[(7-methoxynaphthalen-2-yl)sulfonyl](thiophen-3-ylmethyl)amino}-2-oxopyrrolidin-1-yl]methyl}thiophene-2-carboximidamide, Kallikrein-6 | | Authors: | Chen, X, Zhang, Y, Xia, T, Wang, R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Virtual Screening and X-ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group.
Acs Med.Chem.Lett., 3, 2012
|
|
9GA0
 
 | |
1MH5
 
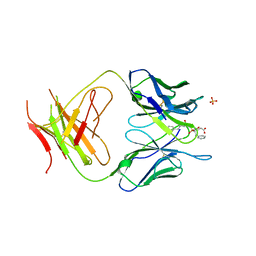 | | The Structure Of The Complex Of The Fab Fragment Of The Esterolytic Antibody MS6-164 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS6-164, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MJN
 
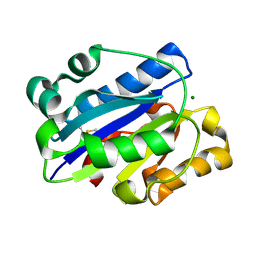 | | Crystal Structure of the intermediate affinity aL I domain mutant | | Descriptor: | Integrin alpha-L, MAGNESIUM ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.H, Yang, Y.T, Dong, Y.C, Jun, C.D, McCormack, A, Zhang, R.G, Wang, J.H, Springer, T.A. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the alphaL I Domain and its Complex with ICAM-1 reveal a Shape-shifting Pathway for Integrin Regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
6ZZJ
 
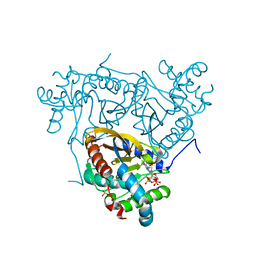 | | Crystal structure of the catalytic domain of Corynebacterium glutamicum acetyltransferase AceF (E2p) in complex with oxidized CoA. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, OXIDIZED COENZYME A | | Authors: | Bruch, E.M, Lexa-Sapart, N, Bellinzoni, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Actinobacteria challenge the paradigm: A unique protein architecture for a well-known, central metabolic complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SFJ
 
 | | ChRmine in MSP1E3D1 lipid nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ChRmine, RETINAL | | Authors: | Tucker, K, Brohawn, S. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of the channelrhodopsin ChRmine in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
6SX2
 
 | | dsRNA recognition by R38AK41A mutant of H7N1 NS1 RNA Binding Domain | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 1, RNA (5'-R(*GP*GP*UP*AP*AP*CP*UP*GP*UP*UP*AP*CP*AP*GP*UP*UP*AP*CP*C)-3') | | Authors: | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
3MZO
 
 | |
9GH9
 
 | |
7SFK
 
 | | ChRmine in MSP1E3D1 lipid nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ChRmine, RETINAL | | Authors: | Tucker, K, Brohawn, S. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of the channelrhodopsin ChRmine in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
9GLY
 
 | |
9GJV
 
 | |
4FWU
 
 | | Crystal structure of glutaminyl cyclase from drosophila melanogaster in space group I4 | | Descriptor: | 1,2-ETHANEDIOL, CG32412, SULFATE ION, ... | | Authors: | Kolenko, P, Koch, B, Stubbs, M.T. | | Deposit date: | 2012-07-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of glutaminyl cyclase from Drosophila melanogaster in space group I4.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
7AAQ
 
 | | sugar/H+ symporter STP10 in outward occluded conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ACETATE ION, ... | | Authors: | Bavnhoej, L, Paulsen, P.A, Pedersen, B.P. | | Deposit date: | 2020-09-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Molecular mechanism of sugar transport in plants unveiled by structures of glucose/H + symporter STP10.
Nat.Plants, 7, 2021
|
|
3N2K
 
 | | TUBULIN-NSC 613862: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Barbier, P, Dorleans, A, Devred, F, Sanz, L, Allegro, D, Alfonso, C, Knossow, M, Peyrot, V, Andreu, J.M. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers.
J.Biol.Chem., 285, 2010
|
|
6SPQ
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
