4DHV
 
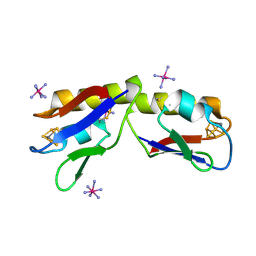 | | Crystal structure of the Pyrococcus furiosus ferredoxin D14C variant containing the heterometallic [AgFe3S4] cluster | | Descriptor: | COBALT HEXAMMINE(III), Ferredoxin, SILVER/IRON/SULFUR CLUSTER | | Authors: | Jakab-Simon, I.N, Christensen, H.E.M, Haahr, L.T. | | Deposit date: | 2012-01-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Heterometallic [AgFe(3)S (4)] ferredoxin variants: synthesis, characterization, and the first crystal structure of an engineered heterometallic iron-sulfur protein.
J.Biol.Inorg.Chem., 18, 2013
|
|
2E8U
 
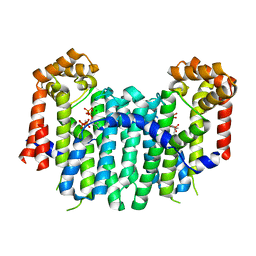 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and IPP (P21) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Chen, C.K.-M, Ko, T.P, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E91
 
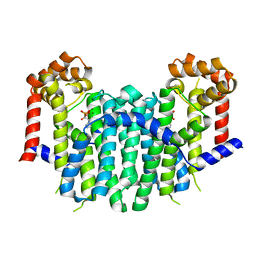 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-91 | | Descriptor: | Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION, ZOLEDRONIC ACID | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Chen, C.K.-M, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4N4U
 
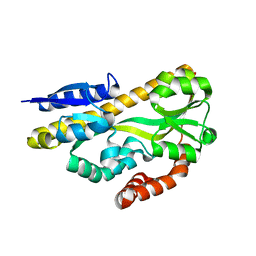 | | Crystal structure of ABC transporter solute binding protein BB0719 from Bordetella bronchiseptica RB50, TARGET EFI-510049 | | Descriptor: | GLYCEROL, Putative ABC transporter periplasmic solute-binding protein | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1V6U
 
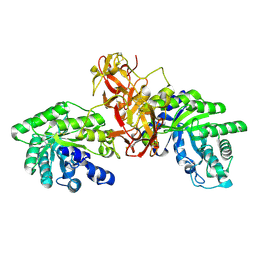 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose | | Descriptor: | alpha-D-xylopyranose, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
4N4W
 
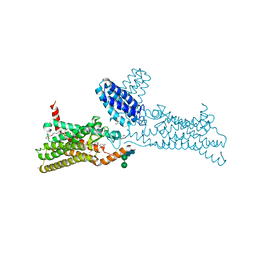 | | Structure of the human smoothened receptor in complex with SANT-1. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (E)-N-(4-benzylpiperazin-1-yl)-1-(3,5-dimethyl-1-phenyl-1H-pyrazol-4-yl)methanimine, Cytochrome b(562),Smoothened homolog, ... | | Authors: | Wang, C, Wu, H, Han, G.W, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
6H8F
 
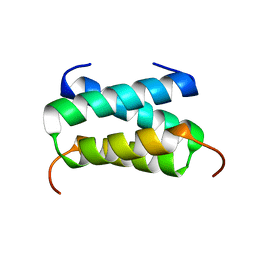 | | Fragment of the C-terminal domain of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | TssA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-08-02 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
1GGU
 
 | | HUMAN FACTOR XIII WITH CALCIUM BOUND IN THE ION SITE | | Descriptor: | CALCIUM ION, PROTEIN (COAGULATION FACTOR XIII) | | Authors: | Fox, B.A, Yee, V.C, Pederson, L.C, Trong, I.L, Bishop, P.D, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 1998-07-22 | | Release date: | 1999-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by x-ray crystallography.
J.Biol.Chem., 274, 1999
|
|
3RN8
 
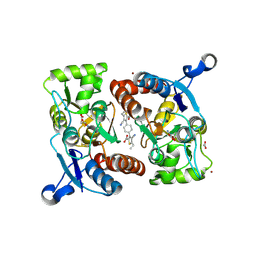 | |
6GM7
 
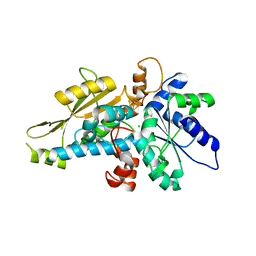 | | [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii,variant E144A | | Descriptor: | CHLORIDE ION, Fe-hydrogenase, IRON/SULFUR CLUSTER | | Authors: | Duan, J, Engelbrecht, V, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
1V6W
 
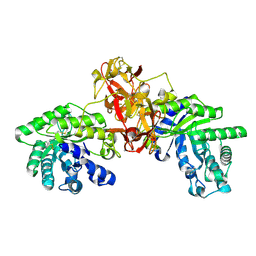 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ENDO-1,4-BETA-D-XYLANASE, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
4EP6
 
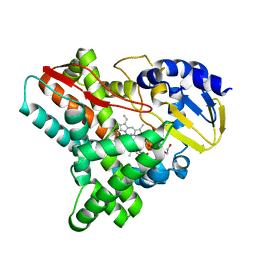 | | Crystal structure of the XplA heme domain in complex with imidazole and PEG | | Descriptor: | Cytochrome P450-like protein XplA, IMIDAZOLE, PENTAETHYLENE GLYCOL, ... | | Authors: | Bui, S.H, McLean, K.J, Cheesman, M.R, Bradley, J.M, Rigby, S.E.J, Leys, D, Munro, A.W. | | Deposit date: | 2012-04-17 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unusual Spectroscopic and Ligand Binding Properties of the Cytochrome P450-Flavodoxin Fusion Enzyme XplA.
J.Biol.Chem., 287, 2012
|
|
2AHO
 
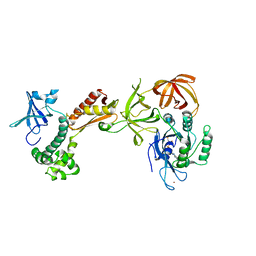 | | Structure of the archaeal initiation factor eIF2 alpha-gamma heterodimer from Sulfolobus solfataricus complexed with GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Translation initiation factor 2 alpha subunit, ... | | Authors: | Yatime, L, Mechulam, Y, Blanquet, S, Schmitt, E. | | Deposit date: | 2005-07-28 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Switch of the gamma Subunit in an Archaeal aIF2alphagamma Heterodimer
Structure, 14, 2006
|
|
3F84
 
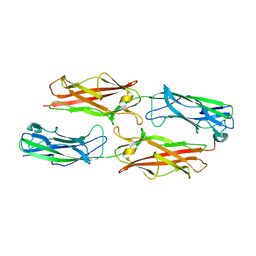 | |
3UAO
 
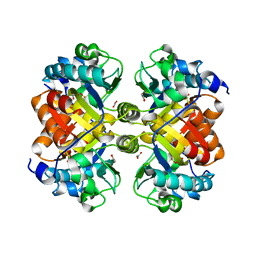 | | Structure and Catalytic Mechanism of the Vitamin B3 Degradative Enzyme Maleamate Amidohydrolase from Bordetalla bronchiseptica RB50 | | Descriptor: | ACETATE ION, Putative isochorismatase | | Authors: | Kincaid, V.A, Sullivan, E.D, Klein, R.D, Rowlett, R.S, Snider, M.J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structure and Catalytic Mechanism of Nicotinate (Vitamin B(3)) Degradative Enzyme Maleamate Amidohydrolase from Bordetella bronchiseptica RB50.
Biochemistry, 51, 2012
|
|
1Z37
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with adenosine | | Descriptor: | ADENOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
3G1F
 
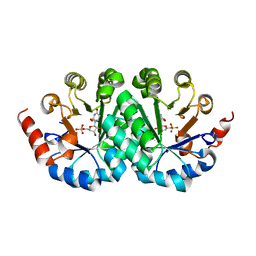 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 5,6-dihydroorotidine 5'-monophosphate | | Descriptor: | 5,6-dihydroorotidine 5'-monophosphate, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1XVU
 
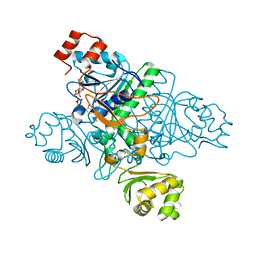 | | Crystal Structure of CaiB mutant D169A in complex with Coenzyme A | | Descriptor: | COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-10-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1XVV
 
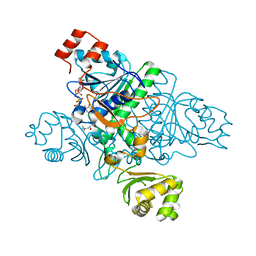 | | Crystal Structure of CaiB mutant D169A in complex with carnitinyl-CoA | | Descriptor: | Crotonobetainyl-CoA:carnitine CoA-transferase, L-CARNITINYL-COA INNER SALT | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-10-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
2CHH
 
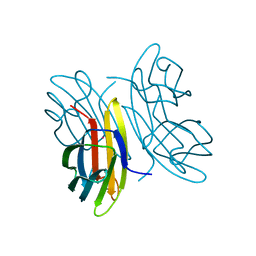 | | RALSTONIA SOLANACEARUM HIGH-AFFINITY MANNOSE-BINDING LECTIN | | Descriptor: | CALCIUM ION, PROTEIN RSC3288, UNKNOWN ATOM OR ION, ... | | Authors: | Mitchell, E.P, Wimmerova, M, Imberty, A. | | Deposit date: | 2006-03-15 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A new Ralstonia solanacearum high-affinity mannose-binding lectin RS-IIL structurally resembling the Pseudomonas aeruginosa fucose-specific lectin PA-IIL.
Mol. Microbiol., 52, 2004
|
|
1SJD
 
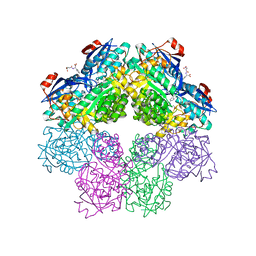 | | x-ray structure of o-succinylbenzoate synthase complexed with n-succinyl phenylglycine | | Descriptor: | N-SUCCINYL PHENYLGLYCINE, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
2CI9
 
 | | Nck1 SH2-domain in complex with a dodecaphosphopeptide from EPEC protein Tir | | Descriptor: | CYTOPLASMIC PROTEIN NCK1, TRANSLOCATED INTIMIN RECEPTOR | | Authors: | Frese, S, Schubert, W.-D, Findeis, A.C, Marquardt, T, Roske, Y.S, Stradal, T.E.B, Heinz, D.W. | | Deposit date: | 2006-03-17 | | Release date: | 2006-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Phosphotyrosine Peptide Binding Specificity of Nck1 and Nck2 Src Homology 2 Domains.
J.Biol.Chem., 281, 2006
|
|
1SJB
 
 | | X-ray structure of o-succinylbenzoate synthase complexed with o-succinylbenzoic acid | | Descriptor: | 2-SUCCINYLBENZOATE, MAGNESIUM ION, N-acylamino acid racemase | | Authors: | Thoden, J.B, Taylor-Ringia, E.A, Garrett, J.B, Gerlt, J.A, Holden, H.M, Rayment, I. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis
Biochemistry, 43, 2004
|
|
3G8R
 
 | | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472 | | Descriptor: | Probable spore coat polysaccharide biosynthesis protein E, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-12 | | Release date: | 2009-04-07 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472
To be Published
|
|
