5UEH
 
 | | Structure of GSTO1 covalently conjugated to quinolinic acid fluorosulfate | | Descriptor: | 2-(4-chlorophenyl)-6-[(fluorosulfonyl)oxy]quinoline-4-carboxylic acid, GLYCEROL, Glutathione S-transferase omega-1, ... | | Authors: | Mortenson, D.E, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
5COD
 
 | | Bovine heart complex I membrane domain | | Descriptor: | NADH-ubiquinone oxidoreductase chain 4, NADH-ubiquinone oxidoreductase chain 5, SDAP, ... | | Authors: | Zhu, J, Hirst, J, King, M.S, Yu, M, Leslie, A.G.W, Klipcan, L. | | Deposit date: | 2015-07-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.74 Å) | | Cite: | Structure of subcomplex I beta of mammalian respiratory complex I leads to new supernumerary subunit assignments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1OFI
 
 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
7MK0
 
 | |
6SL6
 
 | | p53 charged core | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION | | Authors: | Gallardo, R, Langenberg, T, Schymkowitz, J, Rousseau, F, Ulens, C. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Thermodynamic and Evolutionary Coupling between the Native and Amyloid State of Globular Proteins.
Cell Rep, 31, 2020
|
|
6SLB
 
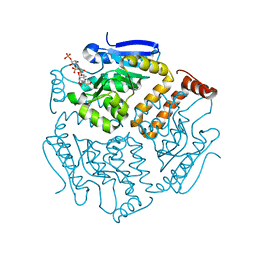 | | Crystal structure of isomerase PaaG with trans-3,4-didehydroadipyl-CoA | | Descriptor: | (~{E})-6-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]-6-oxidanylidene-hex-3-enoic acid, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Saleem-Batcha, R, Spieker, M, Teufel, R. | | Deposit date: | 2019-08-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Mechanistic Basis of an Oxepin-CoA Forming Isomerase in Bacterial Primary and Secondary Metabolism.
Acs Chem.Biol., 14, 2019
|
|
8TDC
 
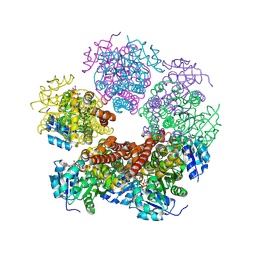 | | Structure of PYCR1 complexed with NADH and 1,3-dithiane-2-carboxylic acid | | Descriptor: | 1,3-dithiane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD9
 
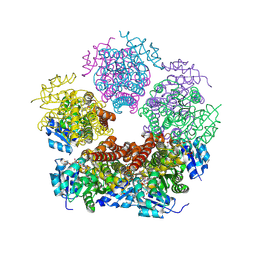 | | Structure of PYCR1 complexed with NADH and L-pipecolic acid | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD3
 
 | |
8TDB
 
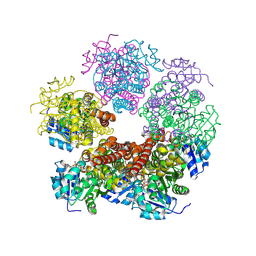 | | Structure of PYCR1 complexed with NADH and 1-hydroxyethane-1-sulfonate | | Descriptor: | (1R)-1-hydroxyethane-1-sulfonic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD4
 
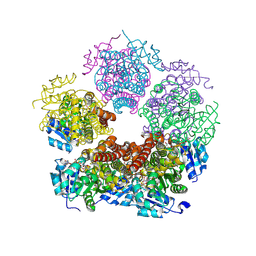 | | Structure of PYCR1 complexed with NADH and 1,3-Dithiolane-2-carboxylic acid | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD2
 
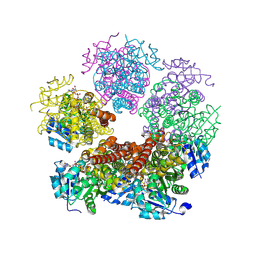 | | Structure of PYCR1 complexed with NADH and cyclobutane-1,1-dicarboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD7
 
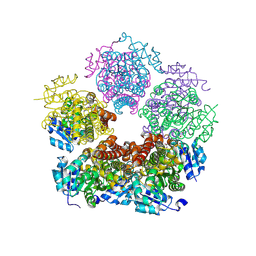 | | Structure of PYCR1 complexed with 2S-hydroxy-3-methylbutyric acid | | Descriptor: | (2S)-2-hydroxy-3-methylbutanoic acid, DI(HYDROXYETHYL)ETHER, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TDD
 
 | | Structure of PYCR1 complexed with NADH and 2-(furan-2-yl)acetic acid | | Descriptor: | (furan-2-yl)acetic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD6
 
 | | Structure of PYCR1 complexed with NADH and 2-(Methylthio)acetic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD8
 
 | | Structure of PYCR1 complexed with NADH and 2S-Hydroxy-3,3-dimethylbutyric acid | | Descriptor: | (2S)-2-hydroxy-3,3-dimethylbutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD5
 
 | | Structure of PYCR1 complexed with NADH and Tetrahydrothiophene-2-carboxylic acid | | Descriptor: | (2R)-thiolane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
6SBI
 
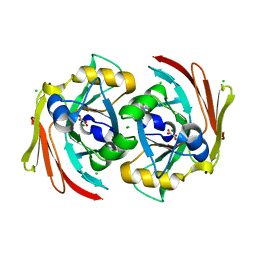 | | X-ray structure of murine Fumarylacetoacetate hydrolase domain containing protein 1 (FAHD1) in complex with inhibitor oxalate | | Descriptor: | Acylpyruvase FAHD1, mitochondrial, CHLORIDE ION, ... | | Authors: | Rupp, B, Naschberger, A, Weiss, A.K.H. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional comparison of fumarylacetoacetate domain containing protein 1 in human and mouse.
Biosci.Rep., 40, 2020
|
|
6SL9
 
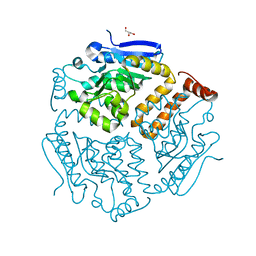 | |
5AK2
 
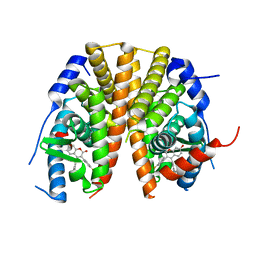 | | Oxyphenylpropenoic acids as Oral Selective Estrogen Receptor Down- Regulators. | | Descriptor: | (E)-3-[4-[[3-(4-fluoranyl-2-methyl-phenyl)-7-oxidanyl-2-oxidanylidene-chromen-4-yl]methyl]phenyl]prop-2-enoic acid, ESTROGEN RECEPTOR | | Authors: | Degorce, S, Bailey, A, Callis, R, De Savi, C, Ducray, R, Lamot, P, MacFaul, P, Maudet, M, Norman, R.A, Scott, J.S, Phillips, C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Investigation of (E)-3-[4-(2-Oxo-3-Aryl-Chromen-4-Yl)Oxyphenyl]Acrylic Acids as Oral Selective Estrogen Receptor Down-Regulators.
J.Med.Chem., 58, 2015
|
|
1OFH
 
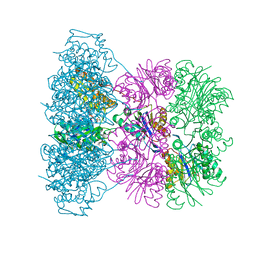 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
6H1B
 
 | | Structure of amide bond synthetase Mcba K483A mutant from Marinactinospora thermotolerans | | Descriptor: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | Authors: | Rowlinson, B, Petchey, M, Cuetos, A, Frese, A, Dannevald, S, Grogan, G. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Broad Aryl Acid Specificity of the Amide Bond Synthetase McbA Suggests Potential for the Biocatalytic Synthesis of Amides.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6SBJ
 
 | | X-ray structure of mus musculus Fumarylacetoacetate hydrolase domain containing protein 1 (FAHD1) apo-form uuncomplexed | | Descriptor: | Acylpyruvase FAHD1, mitochondrial, CHLORIDE ION, ... | | Authors: | Rupp, B, Naschberger, A, Weiss, A.K.H. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional comparison of fumarylacetoacetate domain containing protein 1 in human and mouse.
Biosci.Rep., 40, 2020
|
|
8B9B
 
 | |
8B9C
 
 | | S. cerevisiae pol alpha - replisome complex | | Descriptor: | Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA polymerase alpha catalytic subunit A, ... | | Authors: | Jones, M.L, Yeeles, J.T.P. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | How Pol alpha-primase is targeted to replisomes to prime eukaryotic DNA replication.
Mol.Cell, 83, 2023
|
|
