4OYQ
 
 | | (6-isothiocyanatohexyl)benzene inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | 6-isothiocyanatohexylbenzene, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Vo, C.T, Rutledge, M.T, Nakatani, Y, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | (6-isothiocyanatohexyl)benzene inhibitor complexed with Macrophage Migration Inhibitory Factor
To be published
|
|
7U2H
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, aminoacylated P-site fMet-NH-tRNAmet, and deacylated E-site tRNAgly at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8HTT
 
 | | Cryo-EM structure of human TMEM87A, gluconate-bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-gluconic acid, Transmembrane protein 87A,EGFP, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
9R2M
 
 | | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map) | | Descriptor: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | Authors: | Chakraborty, D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2025-04-30 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map)
To Be Published
|
|
8DQW
 
 | |
5HLP
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH BRD3937 | | Descriptor: | 4-(2-methoxyphenyl)-3,7,7-trimethyl-1,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
8INZ
 
 | | Cryo-EM structure of human HCN3 channel in apo state | | Descriptor: | 4-[[(2~{S},4~{a}~{R},6~{S},8~{a}~{S})-6-[(4~{S},5~{R})-4-[(2~{S})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structure of human HCN3 channel and its regulation by cAMP.
J.Biol.Chem., 300, 2024
|
|
5IGA
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with co-purified parahydroxybenzoate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, P-HYDROXYBENZOIC ACID, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-27 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with co-purified parahydroxybenzoate
To be published
|
|
5IIZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F0 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IZZ
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand | | Descriptor: | 3-HYDROXYPHENYLACETATE, DI(HYDROXYETHYL)ETHER, TRAP TRANSPORTER SOLUTE BINDING PROTEIN | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-03-26 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand
To be published
|
|
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5HLN
 
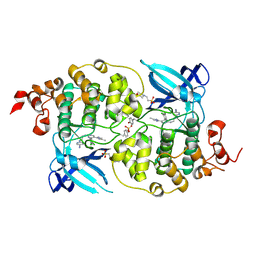 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH CHIR99021 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHIR99021, Glycogen synthase kinase-3 beta, ... | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
8FCY
 
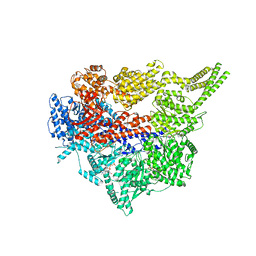 | | Engineered human dynein motor domain in microtubule-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5I7I
 
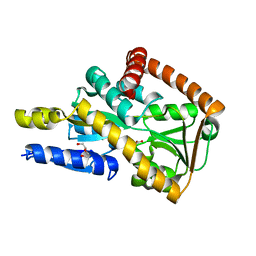 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-crystallized 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, TRAP solute Binding Protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-17 | | Release date: | 2017-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate
To Be Published
|
|
8FD6
 
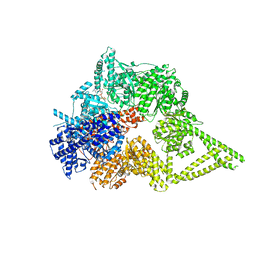 | | Engineered human dynein motor domain in the microtubule-unbound state in the buffer containing ATP-Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, ... | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-02 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8KB4
 
 | | Cryo-EM structure of human TMEM87A A308M | | Descriptor: | (9R,12R)-15-amino-12-hydroxy-6,12-dioxo-7,11,13-trioxa-12lambda~5~-phosphapentadecan-9-yl undecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 87A,EGFP | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
9J8K
 
 | | Crystal structure of the GluA2 ligand binding core (S1S2J) in complex with fluorophore-ligand conjugate | | Descriptor: | 2-(3-azanyl-6-azanylidene-4,5-disulfo-xanthen-9-yl)-5-[[1-[4-(2-hydroxy-2-oxoethyl)-2,3-bis(oxidanylidene)-7-(trifluoromethyl)-1~{H}-quinoxalin-6-yl]pyrrol-3-yl]methylcarbamoyl]benzoic acid, DI(HYDROXYETHYL)ETHER, GluA2 ligand binding core, ... | | Authors: | Fujiwara, T, Adriel, H, Soga, K, Kiyonaka, S, Nango, E. | | Deposit date: | 2024-08-21 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Rapid and reversible fluorescent probe enables repeated snapshot imaging of AMPA receptors during synaptic plasticity.
Sci Adv, 11, 2025
|
|
8DR4
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) without NTD | | Descriptor: | DNA (5'-D(P*AP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR7
 
 | | Open state of RFC:PCNA bound to a nicked dsDNA | | Descriptor: | DNA (26-MER), DNA (5'-D(P*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR3
 
 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR1
 
 | | Consensus closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR5
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
7LGU
 
 | |
8DY7
 
 | |
