2WB1
 
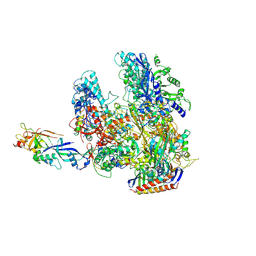 | | The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase | | Descriptor: | DNA-DIRECTED RNA POLYMERASE RPO10 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO11 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO12 SUBUNIT, ... | | Authors: | Korkhin, Y, Unligil, U.M, Littlefield, O, Nelson, P.J, Stuart, D.I, Sigler, P.B, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Evolution of Complex RNA Polymerase: The Complete Archaeal RNA Polymerase Structure
Plos Biol., 7, 2009
|
|
7B9V
 
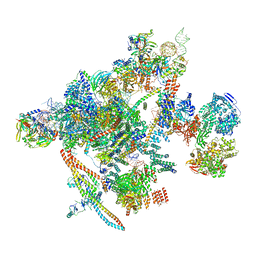 | | Yeast C complex spliceosome at 2.8 Angstrom resolution with Prp18/Slu7 bound | | Descriptor: | 5' exon of UBC4 mRNA, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0054360.mRNA.1.CDS.1, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Nagai, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for conformational equilibrium of the catalytic spliceosome.
Mol.Cell, 81, 2021
|
|
2N55
 
 | |
5GAN
 
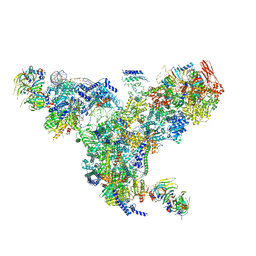 | | The overall structure of the yeast spliceosomal U4/U6.U5 tri-snRNP at 3.7 Angstrom | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-processing factor 31, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
2WTN
 
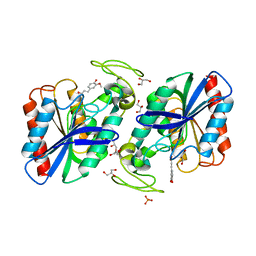 | | Ferulic Acid bound to Est1E from Butyrivibrio proteoclasticus | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, EST1E, GLYCEROL, ... | | Authors: | Goldstone, D.C, Arcus, V.L. | | Deposit date: | 2009-09-17 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of a Promiscuous Feruloyl Esterase (Est1E) from the Rumen Bacterium Butyrivibrio Proteoclasticus.
Proteins, 78, 2010
|
|
5GAP
 
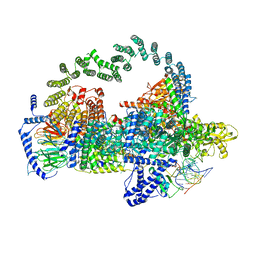 | | Body region of the U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, Pre-mRNA-processing factor 31, Pre-mRNA-splicing factor 6, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Oubridge, C, Bai, X.C, Newman, A, Scheres, S, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
5FKV
 
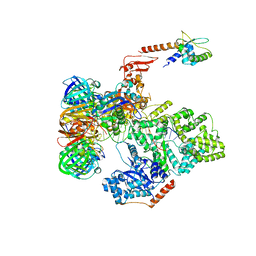 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.04 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
6BK8
 
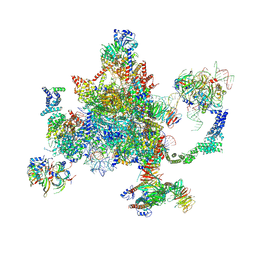 | | S. cerevisiae spliceosomal post-catalytic P complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Lea1, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2017-11-07 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the yeast spliceosomal postcatalytic P complex.
Science, 358, 2017
|
|
2WTM
 
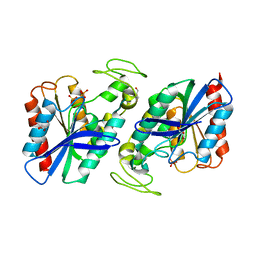 | | Est1E from Butyrivibrio proteoclasticus | | Descriptor: | EST1E, GLYCEROL, PHOSPHATE ION | | Authors: | Goldstone, D.C, Arcus, V.L. | | Deposit date: | 2009-09-17 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Promiscuous Feruloyl Esterase (Est1E) from the Rumen Bacterium Butyrivibrio Proteoclasticus.
Proteins, 78, 2010
|
|
5GM6
 
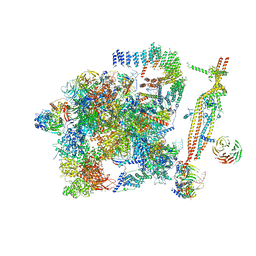 | | Cryo-EM structure of the activated spliceosome (Bact complex) at 3.5 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cold sensitive U2 snRNA suppressor 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a yeast activated spliceosome at 3.5 angstrom resolution
Science, 353, 2016
|
|
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | Descriptor: | RNA polymerase-associated protein RapA, SULFATE ION | | Authors: | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
5GMK
 
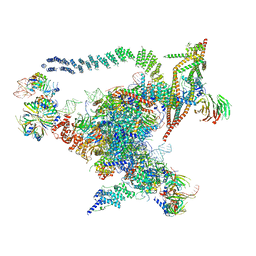 | | Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 3.4 angstrom resolution | | Descriptor: | 5'-Exon, 5'-Splicing Site, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wan, R, Yan, C, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution
Science, 353, 2016
|
|
7BST
 
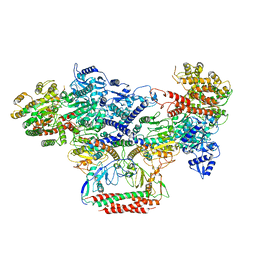 | | EcoR124I-Ocr in the Intermediate State | | Descriptor: | Overcome classical restriction gp0.3, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-03-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.37 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTR
 
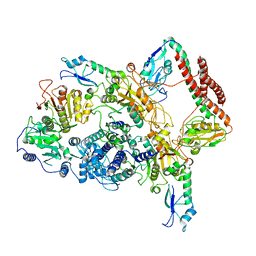 | | EcoR124I-ArdA in the Restriction-Alleviation State | | Descriptor: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
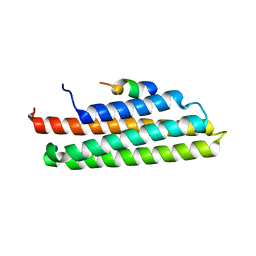
3B71
 
 | |
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
3BG1
 
 | | Architecture of a Coat for the Nuclear Pore Membrane | | Descriptor: | Nucleoporin NUP145, Protein SEC13 homolog | | Authors: | Hoelz, A. | | Deposit date: | 2007-11-23 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architecture of a coat for the nuclear pore membrane.
Cell(Cambridge,Mass.), 131, 2007
|
|
3ASK
 
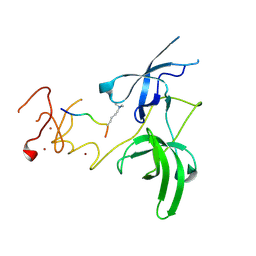 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OE6
 
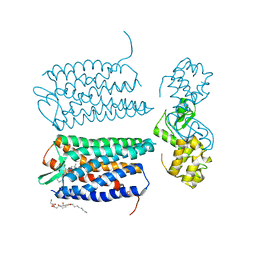 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3OE8
 
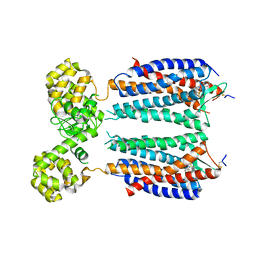 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
3BRV
 
 | | NEMO/IKKb association domain structure | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit beta, NF-kappa-B essential modulator | | Authors: | Silvian, L.F. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a NEMO/IKK-Associating Domain Reveals Architecture of the Interaction Site.
Structure, 16, 2008
|
|
3BG0
 
 | | Architecture of a Coat for the Nuclear Pore Membrane | | Descriptor: | Nucleoporin NUP145, Protein SEC13 homolog | | Authors: | Hoelz, A. | | Deposit date: | 2007-11-23 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Architecture of a coat for the nuclear pore membrane.
Cell(Cambridge,Mass.), 131, 2007
|
|
2PHE
 
 | | Model for VP16 binding to PC4 | | Descriptor: | Alpha trans-inducing protein, TRANSCRIPTIONAL COACTIVATOR PC4 | | Authors: | Jonker, H.R.A, Wechselberger, R.W, Boelens, R, Folkers, G.E, Kaptein, R. | | Deposit date: | 2007-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Properties of the Promiscuous VP16 Activation Domain
Biochemistry, 44, 2005
|
|
