4PRV
 
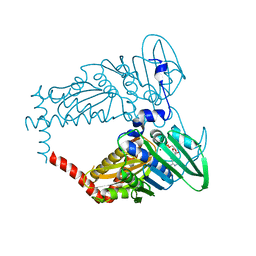 | |
4XTJ
 
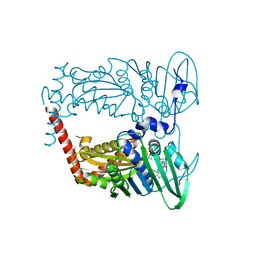 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl plus 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1Y4S
 
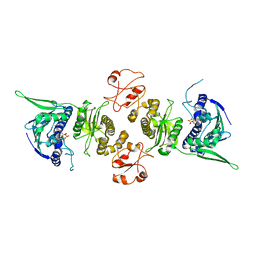 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, MAGNESIUM ION | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
6AZR
 
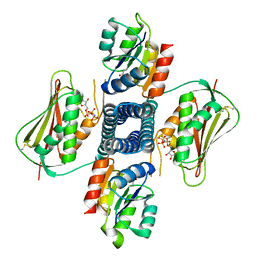 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
5ZXM
 
 | | Crystal Structure of GyraseB N-terminal at 1.93A Resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA gyrase subunit B, ... | | Authors: | Tiwari, P, Gupta, D, Sachdeva, E, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural insights into the transient closed conformation and pH dependent ATPase activity of S.Typhi GyraseB N- terminal domain.
Arch.Biochem.Biophys., 701, 2021
|
|
4ZVI
 
 | | GYRASE B IN COMPLEX WITH 4,5-DIBROMOPYRROLAMIDE-BASED INHIBITOR | | Descriptor: | DNA gyrase subunit B, IODIDE ION, N-(4-{[(4,5-dibromo-1H-pyrrol-2-yl)carbonyl]amino}benzoyl)glycine | | Authors: | Zidar, N, Macut, H, Tomasic, T, Brvar, M, Montalvao, S, Tammela, P, Solmajer, T, Peterlin Masic, L, Ilas, J, Kikelj, D. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Phenyl-4,5-dibromopyrrolamides and N-Phenylindolamides as ATP Competitive DNA Gyrase B Inhibitors: Design, Synthesis, and Evaluation.
J.Med.Chem., 58, 2015
|
|
1Y8O
 
 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y8P
 
 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y8N
 
 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, POTASSIUM ION, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y4U
 
 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
2C2A
 
 | |
2CH4
 
 | |
6DK8
 
 | | RetS kinase region without cobalt | | Descriptor: | NICKEL (II) ION, RetS (Regulator of Exopolysaccharide and Type III Secretion) | | Authors: | Mancl, J.M, Schubot, F.D. | | Deposit date: | 2018-05-29 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Helix Cracking Regulates the Critical Interaction between RetS and GacS in Pseudomonas aeruginosa.
Structure, 27, 2019
|
|
6ENH
 
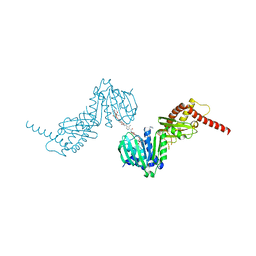 | |
6DK7
 
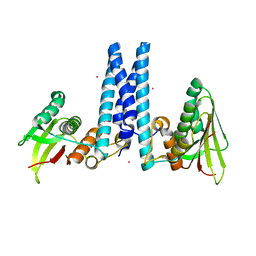 | | RetS histidine kinase region with cobalt | | Descriptor: | COBALT (II) ION, RetS (Regulator of Exopolysaccharide and Type III Secretion) | | Authors: | Mancl, J.M, Schubot, F.D. | | Deposit date: | 2018-05-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Helix Cracking Regulates the Critical Interaction between RetS and GacS in Pseudomonas aeruginosa.
Structure, 27, 2019
|
|
2CG9
 
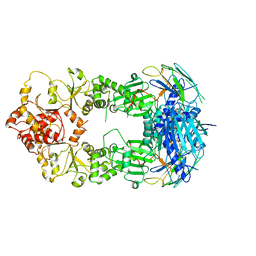 | | Crystal structure of an Hsp90-Sba1 closed chaperone complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, CO-CHAPERONE PROTEIN SBA1 | | Authors: | Ali, M.M.U, Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of an Hsp90-Nucleotide-P23/Sba1 Closed Chaperone Complex
Nature, 440, 2006
|
|
6ENG
 
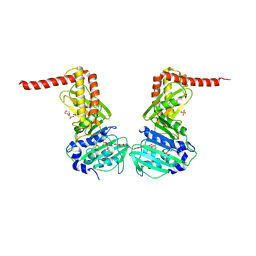 | | Crystal structure of the 43K ATPase domain of Escherichia coli gyrase B in complex with an aminocoumarin | | Descriptor: | CHLORIDE ION, Coumermycin A1, DNA gyrase subunit B, ... | | Authors: | Vanden Broeck, A, McEwen, A.G, Lamour, V. | | Deposit date: | 2017-10-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Gyrase Interaction with Coumermycin A1.
J.Med.Chem., 62, 2019
|
|
4GCZ
 
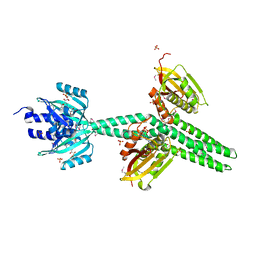 | | Structure of a blue-light photoreceptor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Blue-light photoreceptor, Sensor protein fixL, ... | | Authors: | Diensthuber, R.P, Bommer, M, Moglich, A. | | Deposit date: | 2012-07-31 | | Release date: | 2013-06-19 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators.
Structure, 21, 2013
|
|
4GFH
 
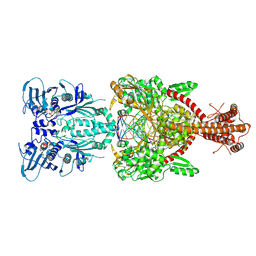 | | Topoisomerase II-DNA-AMPPNP complex | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*AP*CP*TP*GP*CP*TP*AP*C)-3'), ... | | Authors: | Schmidt, B.H, Osheroff, N, Berger, J.M. | | Deposit date: | 2012-08-03 | | Release date: | 2012-10-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (4.408 Å) | | Cite: | Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4I5S
 
 | | Structure and function of sensor histidine kinase | | Descriptor: | Putative histidine kinase CovS; VicK-like protein | | Authors: | Cai, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains
Plos Biol., 11, 2013
|
|
6PAJ
 
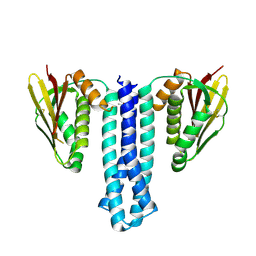 | |
8F71
 
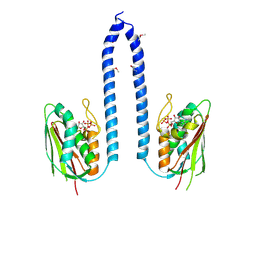 | |
6QRJ
 
 | |
6RGZ
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 6.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH8
 
 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
