2I8A
 
 | |
2I8B
 
 | |
2I8C
 
 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase, MAGNESIUM ION, ... | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I8D
 
 | |
2I8E
 
 | | Structure of SSO1404, a predicted DNA repair-associated protein from Sulfolobus solfataricus P2 | | Descriptor: | Hypothetical protein, IODIDE ION | | Authors: | Wang, S, Zimmerman, M.D, Kudritska, M, Chruszcz, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats.
J.Biol.Chem., 283, 2008
|
|
2I8F
 
 | |
2I8L
 
 | |
2I8N
 
 | |
2I8T
 
 | | GDP-mannose mannosyl hydrolase-calcium-GDP-mannose complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GLYCEROL, ... | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
2I8U
 
 | | GDP-mannose mannosyl hydrolase-calcium-GDP product complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
2I91
 
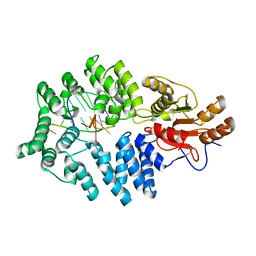 | | 60kDa Ro autoantigen in complex with a fragment of misfolded RNA | | Descriptor: | 5'-R(*C*GP*GP*UP*AP*GP*GP*CP*UP*UP*UP*UP*CP*AP*A)-3', 5'-R(*GP*CP*CP*UP*AP*CP*CP*C)-3', 60 kDa SS-A/Ro ribonucleoprotein, ... | | Authors: | Reinisch, K.M, Stein, A.J. | | Deposit date: | 2006-09-04 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2I94
 
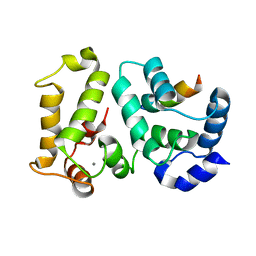 | | NMR Structure of recoverin bound to rhodopsin kinase | | Descriptor: | CALCIUM ION, Recoverin, Rhodopsin kinase | | Authors: | Ames, J.B. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Calcium-induced Inhibition of Rhodopsin Kinase by Recoverin.
J.Biol.Chem., 281, 2006
|
|
2I96
 
 | |
2I99
 
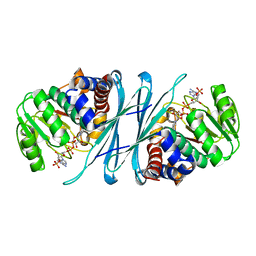 | | Crystal structure of human Mu_crystallin at 2.6 Angstrom | | Descriptor: | Mu-crystallin homolog, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cheng, Z, Sun, L, He, J, Gong, W. | | Deposit date: | 2006-09-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human {micro}-crystallin complexed with NADPH
Protein Sci., 16, 2007
|
|
2I9A
 
 | |
2I9B
 
 | | Crystal structure of ATF-urokinase receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Urokinase plasminogen activator surface receptor, ... | | Authors: | Lubkowski, J, Barinka, C. | | Deposit date: | 2006-09-05 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of interaction between urokinase-type plasminogen activator and its receptor.
J.Mol.Biol., 363, 2006
|
|
2I9C
 
 | | Crystal Structure of the Protein RPA1889 from Rhodopseudomonas palustris CGA009 | | Descriptor: | ACETATE ION, Hypothetical protein RPA1889 | | Authors: | Cymborowski, M.T, Evdokimova, E, Kagan, O, Chruszcz, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Protein RPA1889 from Rhodopseudomonas palustris CGA009
To be Published
|
|
2I9D
 
 | |
2I9E
 
 | | Structure of Triosephosphate Isomerase of Tenebrio molitor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Triosephosphate isomerase | | Authors: | Schmidt, A, Scheerer, P, Wessner, H, Hoehne, W, Krauss, N. | | Deposit date: | 2006-09-05 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A coleopteran triosephosphate isomerase: X-ray structure and phylogenetic impact of insect sequences.
Insect Mol Biol, 19, 2010
|
|
2I9F
 
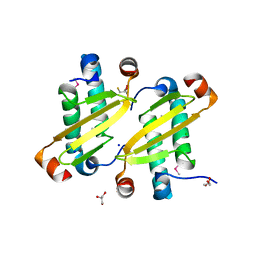 | | Structure of the equine arterivirus nucleocapsid protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Deshpande, A, Wang, S, Walsh, M, Dokland, T. | | Deposit date: | 2006-09-05 | | Release date: | 2007-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the equine arteritis virus nucleocapsid protein reveals a dimer-dimer arrangement.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2I9G
 
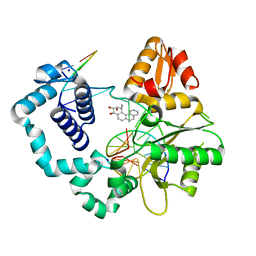 | | DNA Polymerase Beta with a Benzo[c]phenanthrene diol epoxide adducted guanine base | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Wilson, S.H, Beard, W.A, Pedersen, L.C. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of DNA polymerase beta with a benzo[c]phenanthrene diol epoxide-adducted template exhibits mutagenic features.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I9H
 
 | |
2I9I
 
 | | Crystal Structure of Helicobacter pylori protein HP0492 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Mckenzie, C, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-05 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Helicobacter pylori hypothetical protein O25234
TO BE PUBLISHED
|
|
2I9K
 
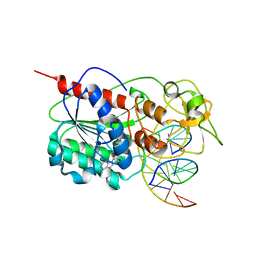 | | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI | | Descriptor: | 5'-D(*T*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3', Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Youngblood, B, Shieh, F.K, De Los Rios, S, Perona, J.J, Reich, N.O. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI
J.Mol.Biol., 362, 2006
|
|
2I9L
 
 | | Structure of Fab 7D11 from a neutralizing antibody against the poxvirus L1 protein | | Descriptor: | Antibody 7D11 heavy chain, Antibody 7D11 light chain, GLYCEROL, ... | | Authors: | Su, H.P, Golden, J.W, Gittis, A.G, Moss, B, Hooper, J.W, Garboczi, D.N. | | Deposit date: | 2006-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the binding of the neutralizing antibody, 7D11, to the poxvirus L1 protein
Virology, 368, 2007
|
|
