4OYQ
 
 | | (6-isothiocyanatohexyl)benzene inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | 6-isothiocyanatohexylbenzene, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Vo, C.T, Rutledge, M.T, Nakatani, Y, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | (6-isothiocyanatohexyl)benzene inhibitor complexed with Macrophage Migration Inhibitory Factor
To be published
|
|
4OSF
 
 | | 4-(2-isothiocyanatoethyl)phenol inhibitor complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Spencer, E.S, Dale, E.J, Gommans, A.L, Vo, C.T, Rutledge, M.T, Nakatani, Y, Gamble, A.B, Smith, R.A.J, Wilbanks, S.M, Hampton, M.B, Tyndall, J.D.A. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Multiple binding modes of isothiocyanates that inhibit macrophage migration inhibitory factor
Eur.J.Med.Chem., 93, 2015
|
|
7STB
 
 | | Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7STE
 
 | | Rad24-RFC ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Checkpoint protein RAD24, ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5I3H
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, POTASSIUM ION, Triosephosphate isomerase, ... | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3F
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3J
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | SODIUM ION, Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3I
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
3EIL
 
 | | Structure of B-DNA d(CGTTAATTAACG)2 in the presence of Manganese | | Descriptor: | 5'-D(*DCP*DGP*DTP*DTP*DAP*DAP*DTP*DTP*DAP*DAP*DCP*DG)-3', MANGANESE (II) ION | | Authors: | Millonig, H, Pous, J, Campos, L.J, Subirana, J.A. | | Deposit date: | 2008-09-16 | | Release date: | 2009-06-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interaction of manganese ions with DNA
J.Inorg.Biochem., 103, 2009
|
|
5I3K
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, SODIUM ION, Triosephosphate isomerase, ... | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
5I3G
 
 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
3FIB
 
 | | RECOMBINANT HUMAN GAMMA-FIBRINOGEN CARBOXYL TERMINAL FRAGMENT (RESIDUES 143-411) BOUND TO CALCIUM AT PH 6.0: A FURTHER REFINEMENT OF PDB ENTRY 1FIB, AND DIFFERS FROM 1FIB BY THE MODELLING OF A CIS PEPTIDE BOND BETWEEN RESIDUES K338 AND C339 | | Descriptor: | CALCIUM ION, FIBRINOGEN GAMMA CHAIN RESIDUES | | Authors: | Pratt, K.P, Cote, H.C.F, Chung, D.W, Stenkamp, R.E, Davie, E.W. | | Deposit date: | 1997-07-14 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
8S3X
 
 | | LIM Domain Kinase 2 (LIMK2) bound to compound 52 | | Descriptor: | 4-(5-cyclopropyl-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)-~{N}-[3-(3-methoxyphenyl)phenyl]-3,6-dihydro-2~{H}-pyridine-1-carboxamide, LIM domain kinase 2 | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Ple, K, Knapp, S. | | Deposit date: | 2024-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Tetrahydropyridine LIMK inhibitors: Structure activity studies and biological characterization.
Eur.J.Med.Chem., 271, 2024
|
|
8INZ
 
 | | Cryo-EM structure of human HCN3 channel in apo state | | Descriptor: | 4-[[(2~{S},4~{a}~{R},6~{S},8~{a}~{S})-6-[(4~{S},5~{R})-4-[(2~{S})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structure of human HCN3 channel and its regulation by cAMP.
J.Biol.Chem., 300, 2024
|
|
8K1M
 
 | | mycobacterial efflux pump, apo state | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2KG4
 
 | | Three-dimensional structure of human Gadd45alpha in solution by NMR | | Descriptor: | Growth arrest and DNA-damage-inducible protein GADD45 alpha | | Authors: | Sanchez, R, Pantoja-Uceda, D, Prieto, J, Diercks, T, Campos-Olivas, R, Blanco, F.J. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase
J.Biol.Chem., 285, 2010
|
|
2LSC
 
 | | Solution structure of 2'F-ANA and ANA self-complementary duplex | | Descriptor: | DNA (5'-D(*(CFL)P*(GFL)P*(CFL)P*(GFL)P*(A5O)P*(A5O)P*(UAR)P*(UAR)P*(CFL)P*(GFL)P*(CFL)P*(GFL))-3') | | Authors: | Martin-Pintado, N, Yahyaee, M, Campos, R, Noronha, A, Wilds, C, Damha, M, Gonzalez, C. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of double helical arabino nucleic acids (ANA and 2'F-ANA): effect of arabinoses in duplex-hairpin interconversion.
Nucleic Acids Res., 40, 2012
|
|
5HLP
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH BRD3937 | | Descriptor: | 4-(2-methoxyphenyl)-3,7,7-trimethyl-1,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
7TVE
 
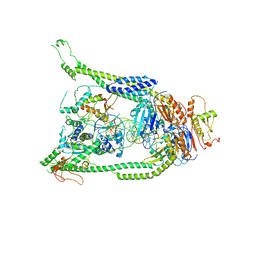 | | ATP and DNA bound SMC5/6 core complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (68-MER), DNA (78-MER), ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of DNA-bound Smc5/6 reveals DNA clamping enabled by multi-subunit conformational changes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5I7I
 
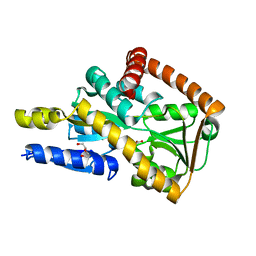 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-crystallized 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, TRAP solute Binding Protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-17 | | Release date: | 2017-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate
To Be Published
|
|
2JHD
 
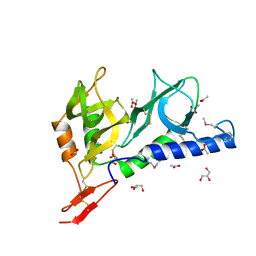 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 3'-sialyl-N-acetyllactosamine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Blumenschein, T.M.A, Friedrich, N, Childs, R.A, Saouros, S, Carpenter, E.P, Campanero-Rhodes, M.A, Simpson, P, Koutroukides, T, Blackman, M.J, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2007-02-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Resolution Insight Into Host Cell Recognition by Toxoplasma Gondii.
Embo J., 26, 2007
|
|
2JH7
 
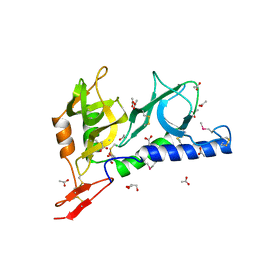 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 6'-sialyl-N-acetyllactosamine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Blumenschein, T.M.A, Friedrich, N, Childs, R.A, Saouros, S, Carpenter, E.P, Campanero-Rhodes, M.A, Simpson, P, Koutroukides, T, Blackman, M.J, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2007-02-20 | | Release date: | 2007-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Atomic Resolution Insight Into Host Cell Recognition by Toxoplasma Gondii.
Embo J., 26, 2007
|
|
5IIZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F0 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IZZ
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand | | Descriptor: | 3-HYDROXYPHENYLACETATE, DI(HYDROXYETHYL)ETHER, TRAP TRANSPORTER SOLUTE BINDING PROTEIN | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-03-26 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157, Triple Surface Mutant K158A_K223A_K313A) in complex with metahydroxyphenylacetate, thermal exchange of ligand
To be published
|
|
