2HAY
 
 | | The Crystal Structure of the Putative NAD(P)H-Flavin Oxidoreductase from Streptococcus pyogenes M1 GAS | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative NAD(P)H-flavin oxidoreductase, SULFATE ION | | Authors: | Kim, Y, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-13 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Crystal Structure of the Putative NAD(P)H-Flavin Oxidoreductase from Streptococcus pyogenes M1 GAS
To be Published, 2006
|
|
2HAZ
 
 | | Crystal structure of the first fibronectin domain of human NCAM1 | | Descriptor: | Neural cell adhesion molecule 1, SODIUM ION | | Authors: | Sekulic, N, Lavie, A. | | Deposit date: | 2006-06-13 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel alpha-helix in the first fibronectin type III repeat of the neural cell adhesion molecule is critical for N-glycan polysialylation.
J.Biol.Chem., 281, 2006
|
|
2HB0
 
 | | Crystal Structure of CfaE, the Adhesive Subunit of CFA/I Fimbria of Enterotoxigenic Escherichia coli | | Descriptor: | CFA/I fimbrial subunit E, DI(HYDROXYETHYL)ETHER, MALONATE ION | | Authors: | Li, Y.F, Xia, D, Poole, S, Rasulova, F, Savarino, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
2HB1
 
 | | Crystal Structure of PTP1B with Monocyclic Thiophene Inhibitor | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wan, Z.-K, Lee, J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monocyclic thiophenes as protein tyrosine phosphatase 1B inhibitors: Capturing interactions with Asp48.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HB2
 
 | | Structure of HIV protease 6X mutant in apo form | | Descriptor: | Protease | | Authors: | Heaslet, H, Tam, K, Elder, J.H, Stout, C.D. | | Deposit date: | 2006-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational flexibility in the flap domains of ligand-free HIV protease.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2HB3
 
 | | Wild-type HIV-1 Protease in complex with potent inhibitor GRL06579 | | Descriptor: | (3AS,5R,6AR)-HEXAHYDRO-2H-CYCLOPENTA[B]FURAN-5-YL (2S,3S)-3-HYDROXY-4-(4-(HYDROXYMETHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design of Novel HIV-1 Protease Inhibitors To Combat Drug Resistance.
J.Med.Chem., 49, 2006
|
|
2HB4
 
 | | Structure of HIV Protease NL4-3 in an Unliganded State | | Descriptor: | MAGNESIUM ION, Protease, R-1,2-PROPANEDIOL | | Authors: | Heaslet, H, Tam, K, Elder, J.H, Stout, C.D. | | Deposit date: | 2006-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational flexibility in the flap domains of ligand-free HIV protease.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2HB5
 
 | | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain | | Descriptor: | MAGNESIUM ION, Reverse transcriptase/ribonuclease H, SULFATE ION | | Authors: | Lim, D, Gregorio, G.G, Bingman, C.A, Martinez-Hackert, E, Hendrickson, W.A, Goff, S.P. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain.
J.Virol., 80, 2006
|
|
2HB6
 
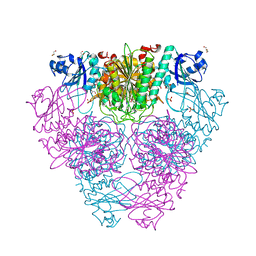 | | Structure of Caenorhabditis elegans leucine aminopeptidase (LAP1) | | Descriptor: | GLYCEROL, Leucine aminopeptidase 1, SODIUM ION, ... | | Authors: | Patskovsky, Y, Zhan, C, Wengerter, B.C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Leucine Aminopeptidase
To be Published
|
|
2HB7
 
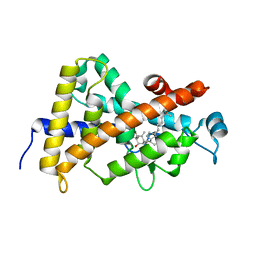 | | Crystal structure of VDR LBD in complex with 2alpha(3-hydroxy-1-propyl) calcitriol | | Descriptor: | 2ALPHA-(3-HYDROXYPROPYL)-1ALPHA,25-DIHYDROXYVITAMIN D3, Vitamin D3 receptor | | Authors: | Hourai, S, Rochel, N, Moras, D. | | Deposit date: | 2006-06-14 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing a Water Channel near the A-Ring of Receptor-Bound 1alpha,25-Dihydroxyvitamin D3 with Selected 2alpha-Substituted Analogues
J.Med.Chem., 49, 2006
|
|
2HB8
 
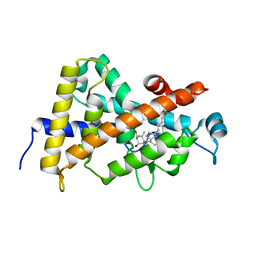 | | Crystal structure of VDR LBD in complex with 2alpha-methyl calcitriol | | Descriptor: | 2ALPHA-METHYL-1ALPHA,25-DIHYDROXY-VITAMIN D3, Vitamin D3 receptor | | Authors: | Hourai, S, Rochel, N, Moras, D. | | Deposit date: | 2006-06-14 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing a Water Channel near the A-Ring of Receptor-Bound 1alpha,25-Dihydroxyvitamin D3 with Selected 2alpha-Substituted Analogues
J.Med.Chem., 49, 2006
|
|
2HB9
 
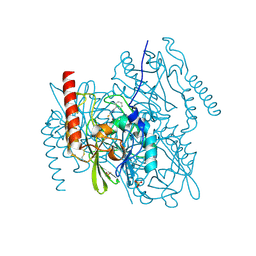 | | Crystal Structure of the Zinc-Beta-Lactamase L1 from Stenotrophomonas Maltophilia (Inhibitor 3) | | Descriptor: | 4-AMINO-5-(2-METHYLPHENYL)-2,4-DIHYDRO-3H-1,2,4-TRIAZOLE-3-THIONE, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
2HBA
 
 | | Crystal Structure of N-terminal Domain of Ribosomal Protein L9 (NTL9) K12M | | Descriptor: | 50S ribosomal protein L9, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Cho, J.-H, Kim, E.Y, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Energetically significant networks of coupled interactions within an unfolded protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2HBB
 
 | | Crystal Structure of the N-terminal Domain of Ribosomal Protein L9 (NTL9) | | Descriptor: | 50S ribosomal protein L9, ZINC ION | | Authors: | Cho, J.-H, Kim, E.Y, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Energetically significant networks of coupled interactions within an unfolded protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2HBC
 
 | | HIGH RESOLUTION X-RAY STRUCTURES OF MYOGLOBIN-AND HEMOGLOBIN-ALKYL ISOCYANIDE COMPLEXES | | Descriptor: | ETHYL ISOCYANIDE, HEMOGLOBIN A (ETHYL ISOCYANIDE) (ALPHA CHAIN), HEMOGLOBIN A (ETHYL ISOCYANIDE) (BETA CHAIN), ... | | Authors: | Johnson, K.A, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1994-08-31 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution X-Ray Structures of Myoglobin-and Hemoglobin-Alkyl Isocyanide Complexes
Thesis, 1, 1993
|
|
2HBD
 
 | | HIGH RESOLUTION X-RAY STRUCTURES OF MYOGLOBIN-AND HEMOGLOBIN-ALKYL ISOCYANIDE COMPLEXES | | Descriptor: | HEMOGLOBIN A (METHYL ISOCYANIDE) (ALPHA CHAIN), HEMOGLOBIN A (METHYL ISOCYANIDE) (BETA CHAIN), METHYL ISOCYANIDE, ... | | Authors: | Johnson, K.A, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1994-08-31 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution X-Ray Structures of Myoglobin-and Hemoglobin-Alkyl Isocyanide Complexes
Thesis, 1, 1993
|
|
2HBE
 
 | | HIGH RESOLUTION X-RAY STRUCTURES OF MYOGLOBIN-AND HEMOGLOBIN-ALKYL ISOCYANIDE COMPLEXES | | Descriptor: | HEMOGLOBIN A (N-BUTYL ISOCYANIDE) (ALPHA CHAIN), HEMOGLOBIN A (N-BUTYL ISOCYANIDE) (BETA CHAIN), N-BUTYL ISOCYANIDE, ... | | Authors: | Johnson, K.A, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1994-08-31 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution X-Ray Structures of Myoglobin-and Hemoglobin-Alkyl Isocyanide Complexes
Thesis, 1, 1993
|
|
2HBF
 
 | | HIGH RESOLUTION X-RAY STRUCTURES OF MYOGLOBIN-AND HEMOGLOBIN-ALKYL ISOCYANIDE COMPLEXES | | Descriptor: | HEMOGLOBIN A (N-PROPYL ISOCYANIDE) (ALPHA CHAIN), HEMOGLOBIN A (N-PROPYL ISOCYANIDE) (BETA CHAIN), N-PROPYL ISOCYANIDE, ... | | Authors: | Johnson, K.A, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1994-08-31 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution X-Ray Structures of Myoglobin-and Hemoglobin-Alkyl Isocyanide Complexes
Thesis, 1, 1993
|
|
2HBG
 
 | |
2HBH
 
 | | Crystal structure of Vitamin D nuclear receptor ligand binding domain bound to a locked side-chain analog of calcitriol and SRC-1 peptide | | Descriptor: | 1,3-CYCLOHEXANEDIOL, 4-METHYLENE-5-[(2E)-[(1S,3AS,7AS)-OCTAHYDRO-1-(5-HYDROXY-5-METHYL-1,3-HEXADIYNYL)-7A-METHYL-4H-INDEN-4-YLIDENE]ETHYLIDENE]-, (1R,3S,5Z), ... | | Authors: | Rochel, N, Hourai, S, Moras, D. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the vitamin D nuclear receptor ligand binding domain in complex with a locked side chain analog of calcitriol
Arch.Biochem.Biophys., 460, 2007
|
|
2HBJ
 
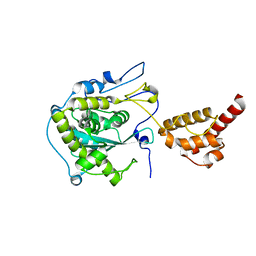 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain | | Descriptor: | Exosome complex exonuclease RRP6 | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HBK
 
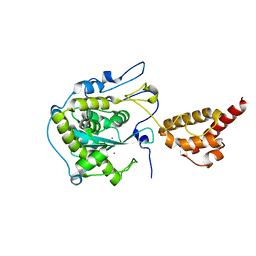 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain; Protein in complex with Mn | | Descriptor: | Exosome complex exonuclease RRP6, MANGANESE (II) ION | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HBL
 
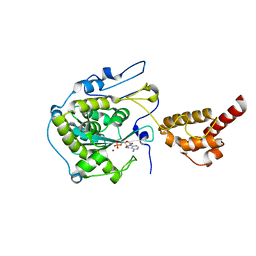 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain; Protein in complex with Mn, Zn, and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Exosome complex exonuclease RRP6, MANGANESE (II) ION, ... | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HBM
 
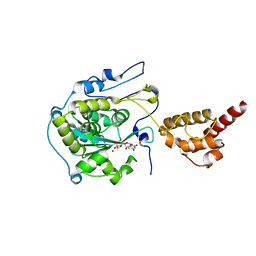 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain; Protein in complex with Mn, Zn, and UMP | | Descriptor: | Exosome complex exonuclease RRP6, MANGANESE (II) ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HBN
 
 | |
