3DFC
 
 | |
2D3B
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with AMPPNP and Methionine sulfoximine | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
5UA3
 
 | | Crystal Structure of a DNA G-Quadruplex with a Cytosine Bulge | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*GP*CP*GP*GP*AP*GP*GP*GP*TP*GP*GP*GP*CP*CP*T)-3'), POTASSIUM ION | | Authors: | Meier, M, McDougall, M.D, Krahn, N.J, Moya-Torres, A, McRae, E.K.S, Booy, E.P, Patel, T.R, McKenna, S.A, Stetefeld, J. | | Deposit date: | 2016-12-18 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8802 Å) | | Cite: | Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge.
Nucleic Acids Res., 46, 2018
|
|
8X3M
 
 | | Crystal structure of p38alpha with an allosteric inhibitor 2 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-(5,6-dimethoxy-1,3-benzothiazol-2-yl)-2-[(4-fluoranylphenoxy)methyl]-1,3-thiazole-4-carboxamide | | Authors: | Hasegawa, S, Kinoshita, T. | | Deposit date: | 2023-11-14 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distinct binding modes of a benzothiazole derivative confer structural bases for increasing ERK2 or p38 alpha MAPK selectivity.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
1UZD
 
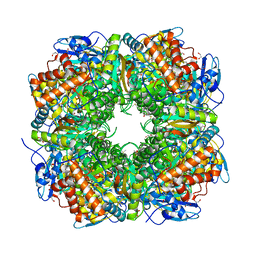 | | Chlamydomonas,Spinach Chimeric Rubisco | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2004-03-11 | | Release date: | 2005-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chimeric Small Subunits Influence Catalysis without Causing Global Conformational Changes in the Crystal Structure of Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase
Biochemistry, 44, 2005
|
|
3E93
 
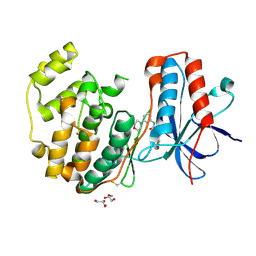 | |
3E92
 
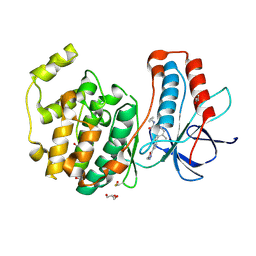 | | Crystal Structure of P38 Kinase in Complex with A Biaryl Amide Inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-cyclopropyl-2',6-dimethyl-4'-(5-methyl-1,3,4-oxadiazol-2-yl)biphenyl-3-carboxamide | | Authors: | Somers, D.O, Patel, S. | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinase array design, back to front: Biaryl amides
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3EH9
 
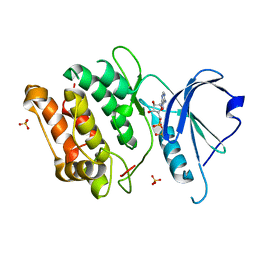 | | Crystal structure of death associated protein kinase complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Death-associated protein kinase 1, SULFATE ION | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
8YD9
 
 | | Crystal structure of p38alpha with an allosteric inhibitor 3 | | Descriptor: | 13-[4-({Imidazo[1,2-a]pyridin-2-yl}methoxy)phenyl]-4,8-dioxa-12,14,16,18-tetraazatetracyclo[9.7.0.0^{3,9}.0^{12,17}]octadeca-1(11),2,9,15,17-pentaen-15-amine, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Hasegawa, S, Kinoshita, T. | | Deposit date: | 2024-02-19 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification of a novel target site for ATP-independent p38 alpha inhibitors
To Be Published
|
|
7ATJ
 
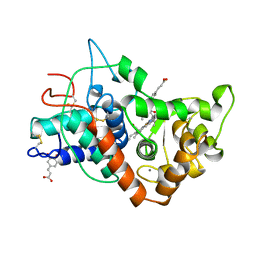 | | RECOMBINANT HORSERADISH PEROXIDASE C1A COMPLEX WITH CYANIDE AND FERULIC ACID | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, CYANIDE ION, ... | | Authors: | Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 1999-04-26 | | Release date: | 2000-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates.
J.Biol.Chem., 274, 1999
|
|
3EDN
 
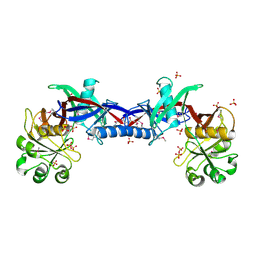 | | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family | | Descriptor: | MAGNESIUM ION, Phenazine biosynthesis protein, PhzF family, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family
To be Published
|
|
3EHA
 
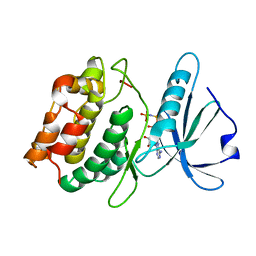 | |
2FQ6
 
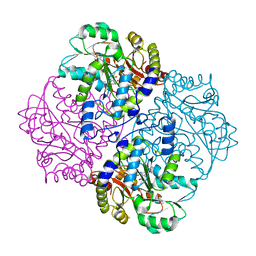 | |
2FSM
 
 | |
2FST
 
 | |
1XA4
 
 | | Crystal structure of CaiB, a type III CoA transferase in carnitine metabolism | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase, ... | | Authors: | Stenmark, P, Gurmu, D, Nordlund, P. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of CaiB, a Type-III CoA Transferase in Carnitine Metabolism
Biochemistry, 43, 2004
|
|
1XB6
 
 | | The K24R mutant of Pseudomonas Aeruginosa Azurin | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Tigerstrom, A, Schwarz, F, Karlsson, G, Okvist, M, Alvarez-Rua, C, Maeder, D, Robb, F.T, Sjolin, L. | | Deposit date: | 2004-08-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Effects of a novel disulfide bond and engineered electrostatic interactions on the thermostability of azurin
Biochemistry, 43, 2004
|
|
2GFS
 
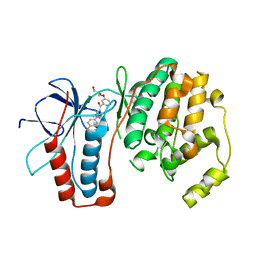 | | P38 Kinase Crystal Structure in complex with RO3201195 | | Descriptor: | Mitogen-Activated Protein Kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Harris, S.F, Bertrand, J, Villasenor, A. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Discovery of S-[5-Amino-1-(4-fluorophenyl)-1H-pyrazol-4-yl]-[3-(2,3-dihydroxypropoxy)phenyl]-methanone (RO3201195), and Orally Bioavailable and Highly Selective Inhibitor of p38 Map Kinase
J.Med.Chem., 49, 2006
|
|
3ETF
 
 | | Crystal structure of a putative succinate-semialdehyde dehydrogenase from salmonella typhimurium lt2 | | Descriptor: | Putative succinate-semialdehyde dehydrogenase | | Authors: | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Wawrzak, Z, Anderson, W.F, Savchenk, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-07 | | Release date: | 2008-11-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
3EFV
 
 | | Crystal Structure of a Putative Succinate-Semialdehyde Dehydrogenase from Salmonella typhimurium LT2 with bound NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative succinate-semialdehyde dehydrogenase | | Authors: | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-10 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
1W5X
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,3-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1VST
 
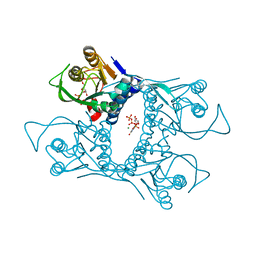 | | Symmetric Sulfolobus solfataricus uracil phosphoribosyltransferase with bound PRPP and GTP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kadziola, A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and kinetic studies of the allosteric transition in Sulfolobus solfataricus uracil phosphoribosyltransferase: Permanent activation by engineering of the C-terminus
J.Mol.Biol., 393, 2009
|
|
3FKN
 
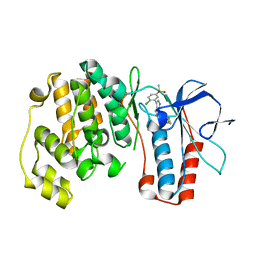 | | P38 kinase crystal structure in complex with RO7125 | | Descriptor: | 3-[2-chloro-5-(methylsulfonyl)phenyl]-6-(2,4-difluorophenoxy)-1H-pyrazolo[3,4-d]pyrimidine, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Ghate, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Binding Pocket Volume: Potential Applications towards Ligand Design and Selectivity
To be Published
|
|
1W07
 
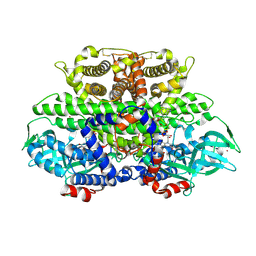 | | Arabidopsis thaliana acyl-CoA oxidase 1 | | Descriptor: | ACYL-COA OXIDASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Henriksen, A, Pedersen, L. | | Deposit date: | 2004-06-01 | | Release date: | 2004-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acyl-Coa Oxidase 1 from Arabidopsis Thaliana. Structure of a Key Enzyme in Plant Lipid Metabolism
J.Mol.Biol., 345, 2005
|
|
3FMH
 
 | |
