7TYP
 
 | | TEAD2 bound to GNE-7883 | | Descriptor: | (8S)-5-(4-cyclohexylphenyl)-3-[3-(fluoromethyl)azetidine-1-carbonyl]-2-(3-methylpyrazin-2-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Fong, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an
expanded Hippo dependent landscape in cancers
To Be Published
|
|
5ZRN
 
 | |
7U37
 
 | |
8OXT
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) H251A VARIANT COMPLEXED WITH N-ACETYLANTHRANILATE AS RESULT OF IN CRYSTALLO TURNOVER OF ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE UNDER HYPEROXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-(ACETYLAMINO)BENZOIC ACID, GLYCEROL, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2023-05-02 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
8D5V
 
 | | WhiB6 bound to the SigmaAr4-RNAP Beta flap tip chimera | | Descriptor: | IRON/SULFUR CLUSTER, Probable transcriptional regulator WhiB6, RNA polymerase sigma factor SigA,DNA-directed RNA polymerase subunit beta | | Authors: | Wan, T, Zhang, L.M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of WhiB6 and SigA4-betaTip complex
To Be Published
|
|
6A0A
 
 | |
8OG0
 
 | |
7UDV
 
 | |
7UDZ
 
 | | Designed pentameric proton channel LQLL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel LQLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J.M, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
8CP2
 
 | |
7UDW
 
 | | Designed pentameric proton channel QQLL | | Descriptor: | De novo designed pentameric proton channel QQLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDY
 
 | | Designed pentameric channel QLLL | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Designed channel QLLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDX
 
 | | Designed pentameric proton channel QLQL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel QLQL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
8DA4
 
 | | Coevolved affibody-Z domain pair LL1.c2 | | Descriptor: | Affibody LL1.FIVM, Immunoglobulin G-binding protein A, SULFATE ION, ... | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8DA7
 
 | | Coevolved affibody-Z domain pair LL1.c6 | | Descriptor: | Immunoglobulin G-binding protein A, MALONATE ION, affibody LL1.FIFV | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
8CQ1
 
 | |
8CPB
 
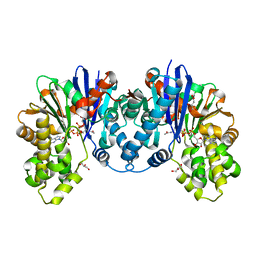 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with AMPPNP, and AnhMurNAc at 1.7 Angstroms resolution. | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, GLYCEROL, ... | | Authors: | Jimenez-Faraco, E, Hermoso, J.A. | | Deposit date: | 2023-03-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
8CP9
 
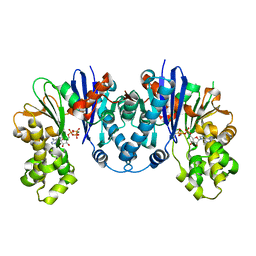 | |
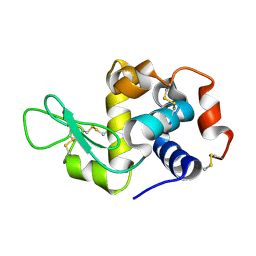
8CPC
 
 | |
5ZZK
 
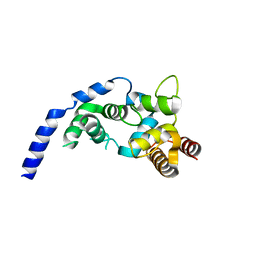 | |
8CQM
 
 | |
8ON5
 
 | |
8P9F
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB161 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[2-(2-azanyl-5-methyl-4-oxidanyl-phenyl)hydrazinyl]benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9L
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB512 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8CZP
 
 | |
