4GS5
 
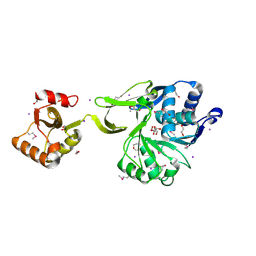 | | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein, IODIDE ION | | Authors: | Tan, K, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-12 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053
To be Published
|
|
1AB9
 
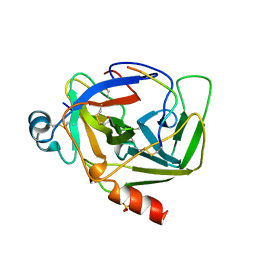 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN | | Descriptor: | GAMMA-CHYMOTRYPSIN, PENTAPEPTIDE (TPGVY), SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-02-05 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
1AFQ
 
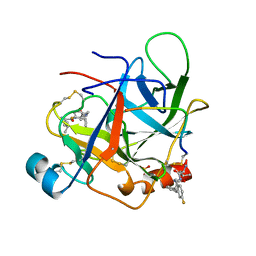 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN COMPLEXED WITH A SYNTHETIC INHIBITOR | | Descriptor: | BOVINE GAMMA-CHYMOTRYPSIN, D-leucyl-N-(4-fluorobenzyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-03-12 | | Release date: | 1997-09-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
5B0N
 
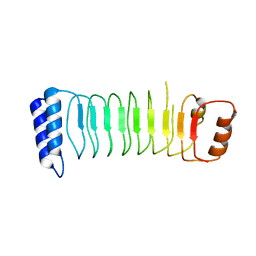 | | Structure of Shigella effector LRR domain | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Takagi, K, Sasakawa, C, Kim, M, Mizushima, T. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the substrate-recognition domain of the Shigella E3 ligase IpaH9.8
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B0T
 
 | | Structure of Shigella effector LRR domain | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Takagi, K, Sasakawa, C, Kim, M, Mizushima, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the substrate-recognition domain of the Shigella E3 ligase IpaH9.8
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1G09
 
 | | CARBONMONOXY LIGANDED BOVINE HEMOGLOBIN PH 7.2 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
3ZPL
 
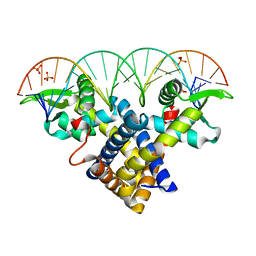 | | Crystal structure of Sco3205, a MarR family transcriptional regulator from Streptomyces coelicolor, in complex with DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*TP*GP*AP*GP*AP*TP*CP*TP *CP*AP*AP*TP*CP*TP*TP*DT)-3', PHOSPHATE ION, PUTATIVE MARR-FAMILY TRANSCRIPTIONAL REPRESSOR | | Authors: | Stevenson, C.E.M, Assaad, A, Lawson, D.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of DNA Sequence Recognition by a Streptomycete Marr Family Transcriptional Regulator Through Surface Plasmon Resonance and X-Ray Crystallography.
Nucleic Acids Res., 41, 2013
|
|
1S0X
 
 | | Crystal structure of the human RORalpha ligand binding domain in complex with cholesterol sulfate at 2.2A | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, Nuclear receptor ROR-alpha | | Authors: | Kallen, J, Schlaeppi, J.M, Bitsch, F, Delhon, I, Fournier, B. | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A
J.Biol.Chem., 279, 2004
|
|
2L0B
 
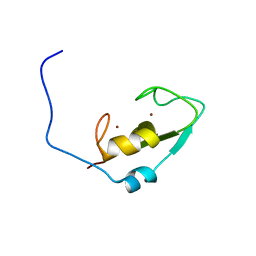 | | Solution NMR structure of zinc finger domain of E3 ubiquitin-protein ligase praja-1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) target HR4710B | | Descriptor: | E3 ubiquitin-protein ligase Praja-1, ZINC ION | | Authors: | Liu, G, Tong, S, Hamilton, K, Ciccosanti, C, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of zinc finger domain of E3 ubiquitin-protein ligase protein praja-1 from Homo sapiens, northeast structural genomics consortium (NESG) target HR4710B
To be Published
|
|
1G08
 
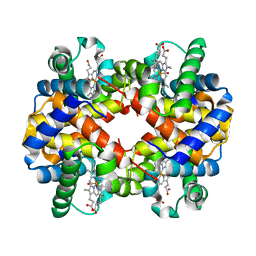 | | CARBONMONOXY LIGANDED BOVINE HEMOGLOBIN PH 5.0 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
3Q10
 
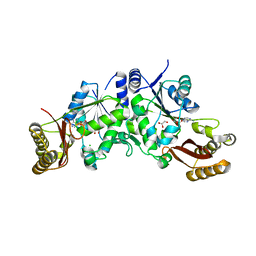 | | Pantoate-beta-alanine ligase from Yersinia pestis | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
1FTL
 
 | |
3RKY
 
 | |
4HV4
 
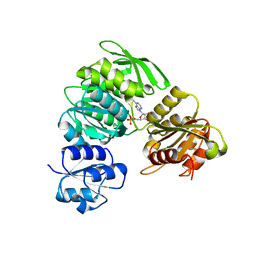 | | 2.25 Angstrom resolution crystal structure of UDP-N-acetylmuramate--L-alanine ligase (murC) from Yersinia pestis CO92 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom resolution crystal structure of UDP-N-acetylmuramate--L-alanine ligase (murC) from Yersinia pestis CO92 in complex with AMP
To be Published
|
|
3RKX
 
 | |
1FTK
 
 | |
1G0A
 
 | | CARBONMONOXY LIGANDED BOVINE HEMOGLOBIN PH 8.5 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
3IAO
 
 | |
3RKW
 
 | |
3RAC
 
 | |
3TUG
 
 | | Crystal structure of the HECT domain of ITCH E3 ubiquitin ligase | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Itchy homolog, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Xue, S, Butler, C, Wernimont, A, Walker, J.R, Tempel, W, Dhe-Paganon, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-16 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the HECT domain of ITCH E3 ubiquitin ligase
To be Published
|
|
6SX3
 
 | | Intercalation of heterocyclic ligand between quartets in G-rich tetrahelical structure | | Descriptor: | VK2, ~{N}2,~{N}6-bis(1-methylquinolin-1-ium-3-yl)pyridine-2,6-dicarboxamide | | Authors: | Kotar, A, Kocman, V, Plavec, J. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Intercalation of a Heterocyclic Ligand between Quartets in a G-Rich Tetrahelical Structure.
Chemistry, 26, 2020
|
|
2LJ6
 
 | |
6O02
 
 | |
2R2X
 
 | | Ricin A-chain (recombinant) complex with Urea | | Descriptor: | Ricin A chain, SULFATE ION, UREA | | Authors: | Carra, J.H, McHugh, C.A, Mulligan, S, Machiesky, L.M, Soares, A.S, Millard, C.B. | | Deposit date: | 2007-08-28 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based identification of determinants of conformational and spectroscopic change at the ricin active site.
Bmc Struct.Biol., 7, 2007
|
|
