6E08
 
 | |
6E07
 
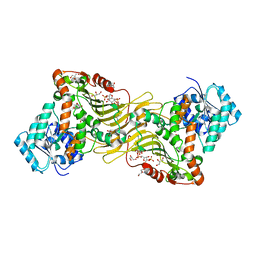 | | Crystal structure of Canton G6PD in complex with structural NADP | | Descriptor: | GLYCEROL, Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Rahighi, S, Mochly-Rosen, D, Wakatsuki, S. | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Correcting glucose-6-phosphate dehydrogenase deficiency with a small-molecule activator.
Nat Commun, 9, 2018
|
|
1LDM
 
 | |
9K72
 
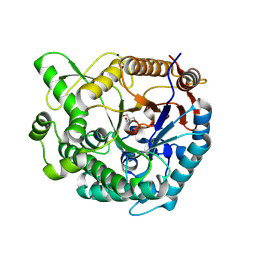 | | Crystal structure of TsaBgl using merged datasets | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-10-23 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Application of Serial Crystallography for Merging Incomplete Macromolecular Crystallography Datasets.
Crystals, 14, 2024
|
|
9K73
 
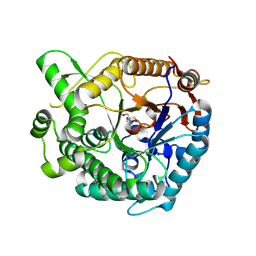 | | Crystal structure of TsaBgl | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | Authors: | Nam, K.H. | | Deposit date: | 2024-10-23 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Application of Serial Crystallography for Merging Incomplete Macromolecular Crystallography Datasets.
Crystals, 14, 2024
|
|
6IN2
 
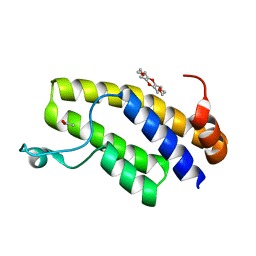 | | Crystal structure of BRD1 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ACETATE ION, Bromodomain-containing protein 1 | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
9JOS
 
 | |
5G4E
 
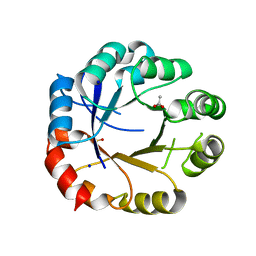 | | S. enterica HisA mutant D10G, Dup13-15, Q24L, G102A, V106L | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, ACETIC ACID, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M, Nasvall, J, Duarte, F, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-05-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ABT
 
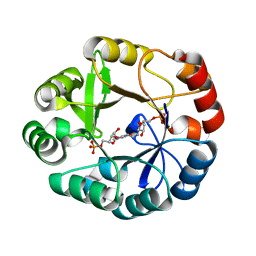 | | S.enterica HisA mutant D7N, G102A, V106M, D176A | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Protein Structures, Functions and Dynamics of Adaptive Evolution
To be Published
|
|
1TNH
 
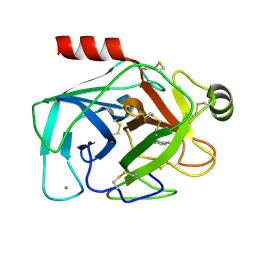 | |
5VZF
 
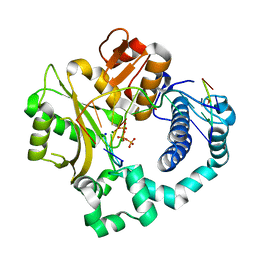 | | Post-catalytic complex of human Polymerase Mu (W434A) mutant with incoming dTTP | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*AP*T)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
1TNJ
 
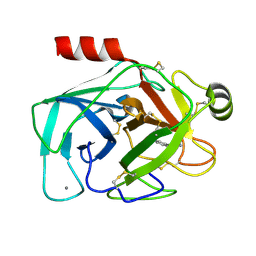 | |
5VZA
 
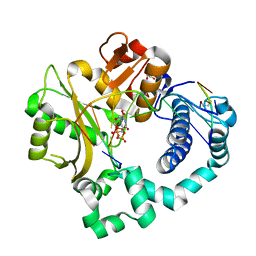 | | Pre-catalytic ternary complex of human Polymerase Mu (G433S) mutant with incoming nonhydrolyzable UMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5VZI
 
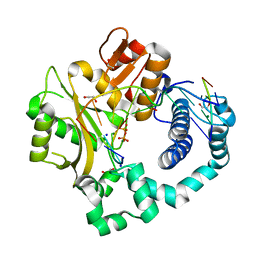 | | Post-catalytic complex of human Polymerase Mu (W434H) mutant with incoming dTTP | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*GP*CP*AP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*AP*T)-3'), ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2017-05-27 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
5FXF
 
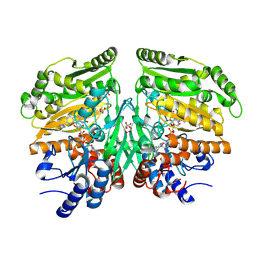 | | Crystal structure of eugenol oxidase in complex with benzoate | | Descriptor: | BENZOIC ACID, EUGENOL OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nguyen, Q.-T, de Gonzalo, G, Binda, C, Martinez, A.R, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-03-01 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biocatalytic Properties and Structural Analysis of Eugenol Oxidase from Rhodococcus Jostii Rha1: A Versatile Oxidative Biocatalyst.
Chembiochem, 17, 2016
|
|
5FXP
 
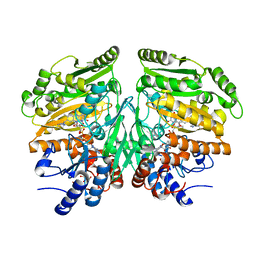 | | Crystal structure of eugenol oxidase in complex with vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, EUGENOL OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nguyen, Q.-T, de Gonzalo, G, Binda, C, Martinez, A.R, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-03-02 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biocatalytic Properties and Structural Analysis of Eugenol Oxidase from Rhodococcus Jostii Rha1: A Versatile Oxidative Biocatalyst.
Chembiochem, 17, 2016
|
|
6FQX
 
 | | Plasmodium falciparum 6-phosphogluconate dehydrogenase in its apo form, in complex with its cofactor NADP+ and in complex with its substrate 6-phosphogluconate | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 6-phosphogluconate dehydrogenase, decarboxylating, ... | | Authors: | Fritz-Wolf, K, Haeussler, K, Reichmann, M, Rahlfs, S, Becker, K. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Plasmodium falciparum 6-Phosphogluconate Dehydrogenase as an Antimalarial Drug Target.
J. Mol. Biol., 430, 2018
|
|
1A6Y
 
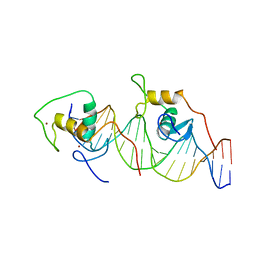 | | REVERBA ORPHAN NUCLEAR RECEPTOR/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*(5IT)P*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*G)-3'), ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Zhao, Q, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 1998-03-04 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elements of an orphan nuclear receptor-DNA complex.
Mol.Cell, 1, 1998
|
|
5H04
 
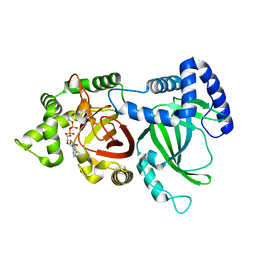 | | Crystal structure of an ADP-ribosylating toxin BECa of a novel binary enterotoxin of C. perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Kawahara, K, Yonogi, S, Munetomo, R, Oki, H, Yoshida, T, Ohkubo, T, Kumeda, Y, Matsuda, S, Kodama, T, Iida, T, Nakamura, S. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Crystal structure of the ADP-ribosylating component of BEC, the binary enterotoxin of Clostridium perfringens.
Biochem.Biophys.Res.Commun., 480, 2016
|
|
5B6K
 
 | |
6MOF
 
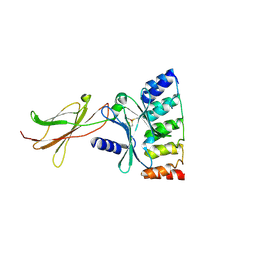 | | Monomeric DARPin G2 complex with EpoR | | Descriptor: | 1,2-ETHANEDIOL, Erythropoietin receptor, G2 DARPin | | Authors: | Jude, K.M, Mohan, K, Garcia, K.C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Topological control of cytokine receptor signaling induces differential effects in hematopoiesis.
Science, 364, 2019
|
|
5YY8
 
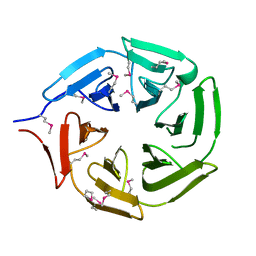 | | Crystal structure of the Kelch domain of human NS1-BP | | Descriptor: | Influenza virus NS1A-binding protein | | Authors: | Guo, L, Liu, Y, Liang, H. | | Deposit date: | 2017-12-08 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Crystal structure of the Kelch domain of human NS1-binding protein at 1.98 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1TNG
 
 | |
4QM1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67 | | Descriptor: | 2-(3-methyl-4-oxo-3,4-dihydrophthalazin-1-yl)-N-(6,7,8,9-tetrahydrodibenzo[b,d]furan-2-yl)acetamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Mandapati, K, Gollapalli, D, Gorla, S.K, Zhang, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-14 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7964 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67
To be Published, 2014
|
|
5YV8
 
 | | Structure of CaMKK2 in complex with CKI-002 | | Descriptor: | 1-amino-4-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
