7RL2
 
 | |
6RMW
 
 | | Structure of N-terminal truncated IMP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
8JUC
 
 | | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening | | Descriptor: | 1,2-ETHANEDIOL, 7-methyl-2-(trifluoromethyl)-3~{H}-[1,2,4]triazolo[1,5-a]pyridin-5-one, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening.
Protein Sci., 33, 2024
|
|
8JOT
 
 | |
5H70
 
 | | Crystal structure of ADP bound dTMP kinase (st1543) from Sulfolobus Tokodaii Strain7 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2016-11-15 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ADP bound dTMP kinase (st1543) from Sulfolobus Tokodaii Strain7
To Be Published
|
|
7RMT
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-70 | | Descriptor: | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
8S63
 
 | | Low pH (5.5) as-isolated MSOX movie series dataset 3 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [1.71 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8S0W
 
 | | High pH (8.0) as-isolated MSOX movie series dataset 1 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [0.35 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8S33
 
 | | Malic semialdehyde dehydrogenase (MSA-DH) from Acinetobacter baumannii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Succinate-semialdehyde dehydrogenase [NAD(P)+] | | Authors: | Piskol, F, Lukat, P, Blankenfeldt, W, Jahn, D, Moser, J. | | Deposit date: | 2024-02-19 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and structural elucidation of the L-carnitine degradation pathway of the human pathogen Acinetobacter baumannii .
Front Microbiol, 15, 2024
|
|
6XUV
 
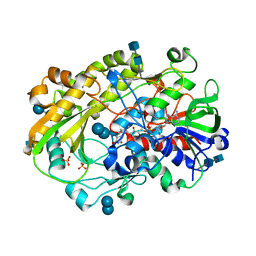 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, laminaribiose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
5KQQ
 
 | |
8BAP
 
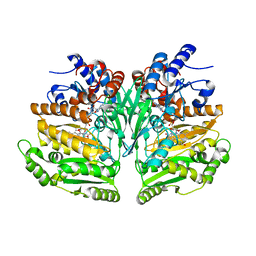 | | Eugenol Oxidase (EUGO) from Rhodococcus jostii RHA1, eightfold mutant active on propanol syringol | | Descriptor: | 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2,6-dimethoxyphenol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | One-Pot Biocatalytic Synthesis of rac -Syringaresinol from a Lignin-Derived Phenol.
Acs Catalysis, 13, 2023
|
|
5NJZ
 
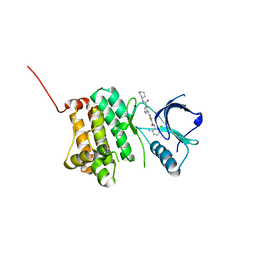 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1g | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-(piperidin-4-ylcarbamoyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NK6
 
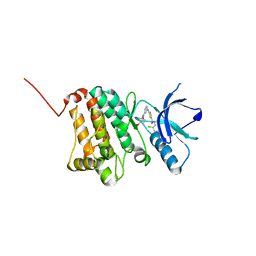 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2d | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[(4-oxidanylcyclohexyl)carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.267 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
9AUX
 
 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
6RME
 
 | | Structure of IMP bound Plasmodium falciparum IMP-nucleotidase mutant D172N | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-08 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
5JE8
 
 | |
8S2R
 
 | | BzdNO in partially coenzyme A - free state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BzdN, BzdO, ... | | Authors: | Ermler, U, Boll, M, Fuchs, J, Demmer, U. | | Deposit date: | 2024-02-19 | | Release date: | 2025-03-05 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymatic Birch reduction via hydrogen atom transfer at [4Fe-4S]-OH 2 and [8Fe-9S] clusters.
Nat Commun, 16, 2025
|
|
9OM9
 
 | |
9OON
 
 | |
9OPG
 
 | |
6FEJ
 
 | | Anabaena Apo-C-Terminal Domain Homolog Protein | | Descriptor: | All4940 protein, UREA | | Authors: | Harris, D, Wilson, A, Muzzopappa, F, Kirilovsky, D, Adir, N. | | Deposit date: | 2018-01-02 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural rearrangements in the C-terminal domain homolog of Orange Carotenoid Protein are crucial for carotenoid transfer.
Commun Biol, 1, 2018
|
|
5MA3
 
 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
7N8F
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988288 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-14 | | Release date: | 2022-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7PUK
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis in complex with Man5 product | | Descriptor: | Beta-N-acetylhexosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
