1OAQ
 
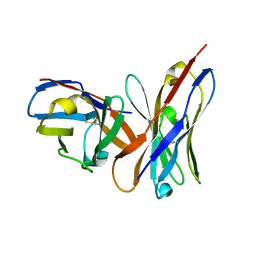 | |
1OF3
 
 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module, TmCBM27 | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
1NT8
 
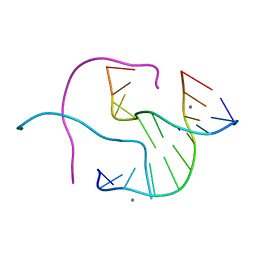 | | Structural Characterisation of the Holliday junction formed by the sequence CCGGTACCGG at 2.00 A | | Descriptor: | 5'-d(CpCpGpGpTpApCpCpGpG)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Texieira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-01-29 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of two Holliday Junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
1OXK
 
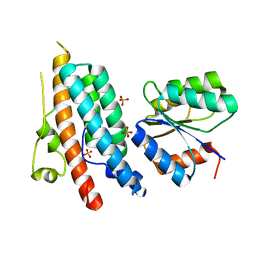 | |
1OYJ
 
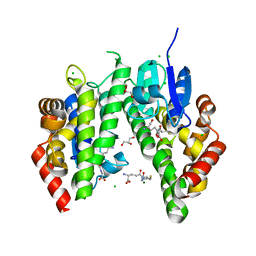 | | Crystal structure solution of Rice GST1 (OsGSTU1) in complex with glutathione. | | Descriptor: | CHLORIDE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Dixon, D.P, McEwen, A.G, Lapthorn, A.J, Edwards, R. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Forced evolution of a herbicide detoxifying glutathione transferase.
J.Biol.Chem., 278, 2003
|
|
1P0A
 
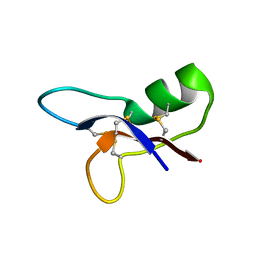 | | NMR structure of ETD135, mutant of the antifungal defensin ARD1 from Archaeoprepona demophon | | Descriptor: | DEFENSIN ARD1 | | Authors: | Landon, C, Guenneugues, M, Barbault, F, Legrain, M, Menin, L, Schott, V, Vovelle, F, Dimarcq, J.L. | | Deposit date: | 2003-04-10 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Lead optimization of antifungal peptides with 3D NMR structures analysis.
Protein Sci., 13, 2004
|
|
1P7K
 
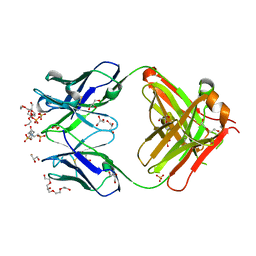 | | Crystal structure of an anti-ssDNA antigen-binding fragment (Fab) bound to 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuermann, J.P, Henzl, M.T, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2003-05-02 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an anti-DNA fab complexed with a non-DNA ligand provides insights into cross-reactivity and molecular mimicry.
Proteins, 57, 2004
|
|
1OZZ
 
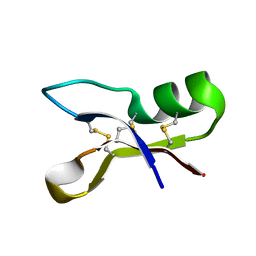 | | NMR structure of antifungal defensin ARD1 from Archaeoprepona demophon | | Descriptor: | defensin ARD1 | | Authors: | Landon, C, Guenneugues, M, Barbault, F, Legrain, M, Menin, L, Schott, V, Vovelle, F, Dimarcq, J.L. | | Deposit date: | 2003-04-10 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Lead optimization of antifungal peptides with 3D NMR structures analysis.
Protein Sci., 13, 2004
|
|
1P00
 
 | | NMR structure of ETD151, mutant of the antifungal defensin ARD1 from Archaeoprepona demophon | | Descriptor: | defensin ARD1 | | Authors: | Landon, C, Guenneugues, M, Barbault, F, Legrain, M, Menin, L, Schott, V, Vovelle, F, Dimarcq, J.L. | | Deposit date: | 2003-04-10 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Lead optimization of antifungal peptides with 3D NMR structures analysis.
Protein Sci., 13, 2004
|
|
1OV3
 
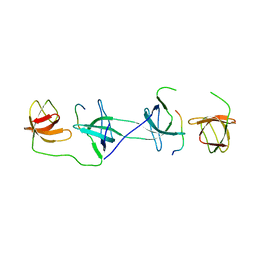 | | Structure of the p22phox-p47phox complex | | Descriptor: | Flavocytochrome b558 alpha polypeptide, Neutrophil cytosol factor 1 | | Authors: | Groemping, Y, Lapouge, K, Smerdon, S.J, Rittinger, K. | | Deposit date: | 2003-03-25 | | Release date: | 2003-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of phosphorylation-induced activation of the NADPH oxidase
Cell(Cambridge,Mass.), 113, 2003
|
|
5B6F
 
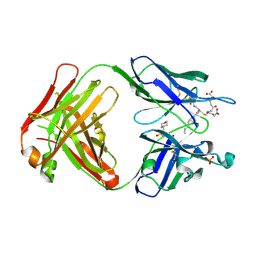 | | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4 | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-(2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4
To Be Published
|
|
5OPO
 
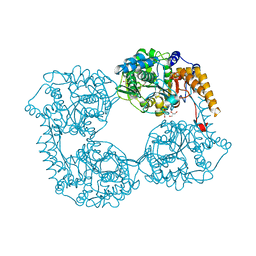 | | Crystal structure of R238G cN-II mutant | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, MAGNESIUM ION | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
6YVZ
 
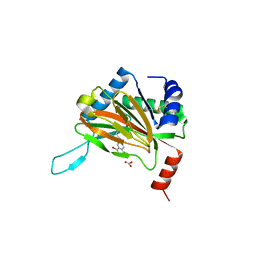 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with bicyclic JLS-367 | | Descriptor: | 4-[(5-bromanylisoquinolin-3-yl)amino]-4-oxidanylidene-butanoic acid, BICARBONATE ION, Egl nine homolog 1, ... | | Authors: | Chowdhury, R, Sorensen, J.L, Schofield, C.J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
6YTW
 
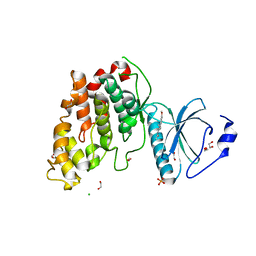 | | CLK3 bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
1ZFE
 
 | | GCA Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*GP*CP*AP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1K54
 
 | | OXA-10 class D beta-lactamase partially acylated with reacted 6beta-(1-hydroxy-1-methylethyl) penicillanic acid | | Descriptor: | (1R)-2-(1-CARBOXY-2-HYDROXY-2-METHYL-PROPYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, 1,2-ETHANEDIOL, Beta lactamase OXA-10, ... | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6YYH
 
 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | Authors: | Lafite, P, Daniellou, R, Bretagne, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
5OQI
 
 | | Crystal Structure of a disulfide trapped single chain trimer composed of the MHC I heavy chain H-2Kb Y84C E63A mutant, beta-2microglobulin, and ovalbumin-derived peptide | | Descriptor: | Beta-2-microglobulin,H-2 class I histocompatibility antigen, K-B alpha chain | | Authors: | Mikolajek, H, Werner, J.M, Beton, M.E. | | Deposit date: | 2017-08-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The partial dissociation of MHC class I-bound peptides exposes their N terminus to trimming by endoplasmic reticulum aminopeptidase 1.
J. Biol. Chem., 293, 2018
|
|
1K7K
 
 | | crystal structure of RdgB- inosine triphosphate pyrophosphatase from E. coli | | Descriptor: | Hypothetical protein yggV | | Authors: | Sanishvili, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
6YVX
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with bicyclic BB-287 | | Descriptor: | 4-(isoquinolin-3-ylamino)-4-oxobutanoic acid, BICARBONATE ION, Egl nine homolog 1, ... | | Authors: | Chowdhury, R, Banerji, B, Schofield, C.J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
1U78
 
 | |
5B7E
 
 | | Structure of perdeuterated CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Akter, M, Higuchi, Y, Shibata, N. | | Deposit date: | 2016-06-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1K9F
 
 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with aldotetraouronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
5AQ8
 
 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, OFF7_DB12V4, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
1ZMD
 
 | | Crystal Structure of Human dihydrolipoamide dehydrogenase complexed to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of Human Dihydrolipoamide Dehydrogenase: NAD+/NADH Binding and the Structural Basis of Disease-causing Mutations
J.Mol.Biol., 350, 2005
|
|
