4CCY
 
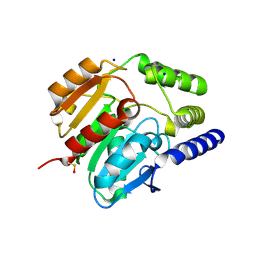 | | Crystal structure of carboxylesterase CesB (YbfK) from Bacillus subtilis | | Descriptor: | CARBOXYLESTERASE YBFK, SODIUM ION | | Authors: | Rozeboom, H.J, Godinho, L.F, Nardini, M, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structures of Two Bacillus Carboxylesterases with Different Enantioselectivities.
Biochim.Biophys.Acta, 1844, 2014
|
|
3EPT
 
 | |
1INY
 
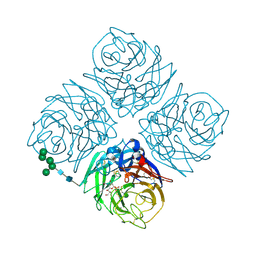 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
2ONJ
 
 | |
4JMY
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with DDIVPC peptide | | Descriptor: | HCV non-structural protein 4A, Polyprotein, SODIUM ION, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2013-03-14 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Importance of the peptide scaffold of drugs that target the hepatitis C virus NS3 protease and its crucial bioactive conformation and dynamic factors.
To be Published
|
|
3G6N
 
 | | Crystal structure of an EfPDF complex with Met-Ala-Ser | | Descriptor: | FE (III) ION, Peptide deformylase, SODIUM ION, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-02-07 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an EfPDF complex with Met-Ala-Ser based on crystallographic packing.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
1JEE
 
 | | Crystal Structure of ATP Sulfurylase in complex with chlorate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
2RTB
 
 | | APOSTREPTAVIDIN, PH 3.32, SPACE GROUP I222 | | Descriptor: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2Q68
 
 | | Crystal Structure of Nak channel D66A, S70E double mutants | | Descriptor: | CALCIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Alam, A, Shi, N, Jiang, Y. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into Ca2+ specificity in tetrameric cation channels.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5O
 
 | | Crystal structure of the C-terminal domain of wzt | | Descriptor: | CHLORIDE ION, Putative ATP binding component of ABC-transporter, SODIUM ION, ... | | Authors: | Kimber, M.S, Cuthbertson, L, Whitfield, C. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Substrate binding by a bacterial ABC transporter involved in polysaccharide export.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3PAK
 
 | | Crystal Structure of Rat Surfactant Protein A neck and carbohydrate recognition domain (NCRD) complexed with Mannose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A, SODIUM ION, ... | | Authors: | Shang, F, Rynkiewicz, M.J, McCormack, F.X, Wu, H, Cafarella, T.M, Head, J, Seaton, B.A. | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change.
J.Biol.Chem., 286, 2011
|
|
3GBV
 
 | | Crystal structure of a putative LacI transcriptional regulator from Bacteroides fragilis | | Descriptor: | 1,2-ETHANEDIOL, Putative LacI-family transcriptional regulator, SODIUM ION | | Authors: | Syed Ibrahim, B, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-10 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative LacI transcriptional regulator from Bacteroides fragilis
To be Published
|
|
4JVM
 
 | | Crystal structure of human estrogen sulfotransferase (SULT1E1) in complex with inactive cofactor PAP and brominated flame retardant TBBPA (tetrabromobisphenol A) | | Descriptor: | 1,2-ETHANEDIOL, 4,4'-propane-2,2-diylbis(2,6-dibromophenol), ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Gosavi, R.A, Knudsen, G.A, Birnbaum, L.S, Pedersen, L.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Mimicking of Estradiol Binding by Flame Retardants and Their Metabolites: A Crystallographic Analysis.
Environ.Health Perspect., 121, 2013
|
|
2XO2
 
 | | Human Annexin V with incorporated Methionine analogue Azidohomoalanine | | Descriptor: | ANNEXIN A5, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Debela, M, Merkel, L, Goettig, P, Budisa, N. | | Deposit date: | 2010-08-09 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Non-Canonical Amino Acids for Click Chemistry Reactions Incorporated in Human Annexin V
To be Published
|
|
3DJ0
 
 | |
4CSH
 
 | | Native structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
4H8R
 
 | | Imipenem complex of GES-5 carbapenemase | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Extended-spectrum beta-lactamase GES-5, IODIDE ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2012-09-23 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases.
J.Am.Chem.Soc., 134, 2012
|
|
4JVL
 
 | | Crystal structure of human estrogen sulfotransferase (SULT1E1) in complex with inactive cofactor PAP and estradiol (E2) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3'-5'-DIPHOSPHATE, ESTRADIOL, ... | | Authors: | Gosavi, R.A, Knudsen, G.A, Birnbaum, L.S, Pedersen, L.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Mimicking of Estradiol Binding by Flame Retardants and Their Metabolites: A Crystallographic Analysis.
Environ.Health Perspect., 121, 2013
|
|
4J6W
 
 | | Crystal structure of HFQ from Pseudomonas aeruginosa in complex with CTP | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Nikulin, A.D, Murina, V, Lekontseva, N. | | Deposit date: | 2013-02-12 | | Release date: | 2013-07-31 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hfq binds ribonucleotides in three different RNA-binding sites.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3H2Z
 
 | |
4C3V
 
 | | Extensive counter-ion interactions with subtilisin in aqueous medium, no Cs soak | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN CARLSBERG, ... | | Authors: | Cianci, M, Negroni, J, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Extensive Counter-Ion Interactions Seen at the Surface of Subtilisin in an Aqueous Medium
Rsc Adv, 2014
|
|
2O11
 
 | |
4JHG
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with trans-zeatin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, MALONATE ION, MtN13 protein, ... | | Authors: | Ruszkowski, M, Tusnio, K, Ciesielska, A, Brzezinski, K, Dauter, M, Dauter, Z, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3CKI
 
 | | Crystal structure of the TACE-N-TIMP-3 complex | | Descriptor: | ADAM 17, Metalloproteinase inhibitor 3, SODIUM ION, ... | | Authors: | Wisniewska, M, Goettig, P, Maskos, K, Belouski, E, Winters, D, Hecht, R, Black, R, Bode, W. | | Deposit date: | 2008-03-15 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE-N-TIMP-3 complex.
J.Mol.Biol., 381, 2008
|
|
1XSL
 
 | | Crystal Structure of human DNA polymerase lambda in complex with a one nucleotide DNA gap | | Descriptor: | 5'-D(*CP*GP*GP*CP*AP*GP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
