9NSW
 
 | | apo-OXA-23, pH 7.5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase OXA-23, SULFATE ION | | Authors: | Smith, C.A, Toth, M, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-03-17 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dual mechanism of the OXA-23 carbapenemase inhibition by the carbapenem NA-1-157.
Antimicrob.Agents Chemother., 69, 2025
|
|
9NT3
 
 | |
9NT6
 
 | | Crystal structure of ecLacI transcription factor ancestor 4 bound to Beta-methyl-D-galactoside | | Descriptor: | 1,2-ETHANEDIOL, LacI Transcription Factor, methyl beta-D-galactopyranoside | | Authors: | Georgelin, R.L, Frkic, R.L, Tan, L, Kaczmarski, J, Jackson, C.J. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of ecLacI transcription factor ancestor 4 bound to Beta-methyl-D-galactoside
To Be Published
|
|
6NI1
 
 | | Crystal Structure of the Beta Lactamase Class A penP from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class A penP from Bacillus subtilis
To Be Published
|
|
6BY4
 
 | | Single-State 14-mer UUCG Tetraloop calculated from Exact NOEs | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
9PGQ
 
 | |
9UGP
 
 | | Crystal structure of MCL-1 in complex with HRK BH3 | | Descriptor: | Activator of apoptosis harakiri, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Wei, H, Wang, J. | | Deposit date: | 2025-04-13 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural analysis of apoptotic MCL1-HRK complex.
Biochem.Biophys.Res.Commun., 768, 2025
|
|
9O63
 
 | | Crystal structure of PLK4 and RP1664 complex | | Descriptor: | 5-cyclopropyl-N~2~-[2,6-difluoro-4-(methanesulfonyl)phenyl]-N~2~-methyl-6-(1-methyl-1H-imidazol-4-yl)-N~4~-(5-methyl-1H-pyrazol-3-yl)pyrimidine-2,4-diamine, SULFATE ION, Serine/threonine-protein kinase PLK4 | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Maderova, Z, Zimmermann, M, Sicheri, F. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of RP-1664: A First-in-Class Orally Bioavailable, Selective PLK4 Inhibitor.
J.Med.Chem., 68, 2025
|
|
9QDT
 
 | | Cytochrome c peroxidase YhjA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Einsle, O, Wuest, A. | | Deposit date: | 2025-03-06 | | Release date: | 2025-07-30 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.572 Å) | | Cite: | Escherichia coli Triheme Enzyme YhjA: Structure and Reactivity.
Biochemistry, 64, 2025
|
|
6NEK
 
 | |
2TOB
 
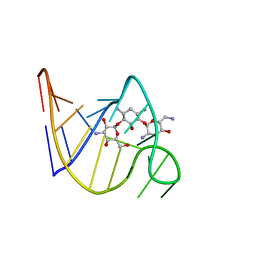 | | SOLUTION STRUCTURE OF THE TOBRAMYCIN-RNA APTAMER COMPLEX, NMR, 13 STRUCTURES | | Descriptor: | 1,3-DIAMINO-4,5,6-TRIHYDROXY-CYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Patel, D.J. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tobramycin-RNA aptamer complex.
Nat.Struct.Biol., 5, 1998
|
|
143D
 
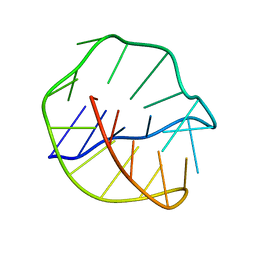 | |
9OOY
 
 | |
9QZE
 
 | |
9O8W
 
 | | Crystal structure of an MKP5 mutant, Y435F, in complex with an allosteric inhibitor | | Descriptor: | 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, Dual specificity protein phosphatase 10, SULFATE ION | | Authors: | Manjula, R, Bennett, A.M, Lolis, E. | | Deposit date: | 2025-04-16 | | Release date: | 2025-07-30 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Dynamic and structural insights into allosteric regulation on MKP5 a dual-specificity phosphatase.
Nat Commun, 16, 2025
|
|
9VFJ
 
 | | PSI-LHCI of Euglena gracilis strain Z | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nakajima, Y, Shen, J.R, Nagao, R. | | Deposit date: | 2025-06-11 | | Release date: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of a divergent photosystem I supercomplex from Euglena gracilis outside green and red lineages
To Be Published
|
|
8VQV
 
 | | Structure of S. odontolytica ZTP riboswitch bound to m-1-pyridinyl-AICA | | Descriptor: | 5-amino-1-(pyridin-3-yl)-1H-imidazole-4-carboxamide, MAGNESIUM ION, RNA (64-MER) | | Authors: | Jones, C.P, Ferre D'Amare, A.R. | | Deposit date: | 2024-01-19 | | Release date: | 2025-06-11 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Machine Learning-Augmented Molecular Dynamics Simulations (MD) Reveal Insights Into the Disconnect Between Affinity and Activation of ZTP Riboswitch Ligands.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
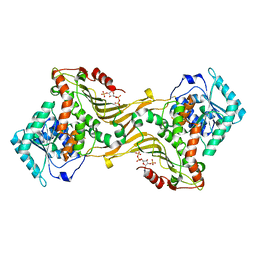
8U2W
 
 | |
4CAT
 
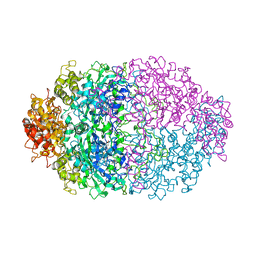 | | THREE-DIMENSIONAL STRUCTURE OF CATALASE FROM PENICILLIUM VITALE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Melik-Adamyan, W.R, Barynin, V.V, Vagin, A.A, Grebenko, A.I. | | Deposit date: | 1983-02-24 | | Release date: | 1983-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of catalase from Penicillium vitale at 2.0 A resolution.
J.Mol.Biol., 188, 1986
|
|
9NSB
 
 | | Crystal structure of an MKP5 allosteric loop mutant, S446G, in complex with an allosteric inhibitor | | Descriptor: | 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, Dual specificity protein phosphatase 10 | | Authors: | Manjula, R, Bennett, A.M, Lolis, E. | | Deposit date: | 2025-03-16 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an MKP5 allosteric loop mutant, S446G, in complex with an allosteric inhibitor
To Be Published
|
|
9RF5
 
 | | M. tuberculosis meets European Lead Factory: identification and structural characterization of novel Rv0183 inhibitors using X-ray crystallography: ELF8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Riegler-Berket, L, Goedl, L, Oberer, M, Polidori, N. | | Deposit date: | 2025-06-04 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | M. tuberculosis meets European Lead Factory: identification and structural characterization of novel Rv0183 inhibitors using X-ray crystallography
To Be Published
|
|
9Q7X
 
 | |
9OMC
 
 | | Crystal structure of E. coli ApaH in complex with Gp4G | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]guanosine, Bis(5'-nucleosyl)-tetraphosphatase [symmetrical], ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2025-05-13 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | ApaH decaps Np 4 N-capped RNAs in two alternative orientations.
Nat.Chem.Biol., 2025
|
|
9OM9
 
 | |
5Z24
 
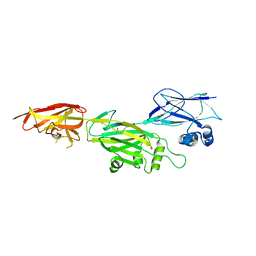 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG - K365A mutant | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-12-28 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
