8FG7
 
 | |
6F4T
 
 | | Human JMJD5 (W414C) in complex with Mn(II), NOG and RCCD1 (139-143) (complex-5) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, JmjC domain-containing protein 5, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2017-11-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
7O28
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 19 (ADO_AE_009) | | Descriptor: | 9-(2-chloranyl-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)-4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 19 (ADO_AE_009)
To Be Published
|
|
4MJO
 
 | | Human liver fructose-1,6-bisphosphatase(d-fructose-1,6-bisphosphate, 1-phosphohydrolase) (e.c.3.1.3.11) complexed with the allosteric inhibitor 3 | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-({4-bromo-6-[(methylcarbamoyl)amino]pyridin-2-yl}carbamoyl)-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Tetaz, T, Benz, J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of protein-ligand binding constants of a cooperatively regulated tetrameric enzyme using electrospray mass spectrometry.
Acs Chem.Biol., 9, 2014
|
|
7O0P
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 10 (ADO_AD_022) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the human METTL3-METTL14 complex bound to Compound 10 (ADO_AD_022)
To Be Published
|
|
6F5W
 
 | | Photorhabdus asymbiotica lectin (PHL) in complex with propargyl-fucoside | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Houser, J, Jancarikova, G, Wimmerova, M, Csavas, M, Borbas, A, Herczeg, M, Fujdiarova, E, Kover, E.K. | | Deposit date: | 2017-12-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Synthesis of alpha-l-Fucopyranoside-Presenting Glycoclusters and Investigation of Their Interaction with Photorhabdus asymbiotica Lectin (PHL).
Chemistry, 24, 2018
|
|
8FG5
 
 | | Apo mouse acidic mammalian chitinase, catalytic domain at 100 K | | Descriptor: | Acidic mammalian chitinase, MAGNESIUM ION | | Authors: | Diaz, R.E, Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2022-12-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
8RYK
 
 | |
8FIU
 
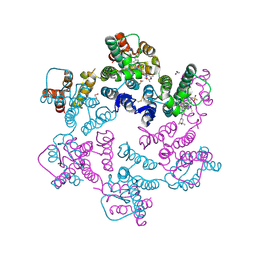 | | Potent long-acting inhibitors targeting HIV-1 capsid based on a versatile quinazolin-4-one scaffold | | Descriptor: | 1,2-ETHANEDIOL, HIV-1 capsid, N-[(1S)-1-{(3P)-3-{4-chloro-3-[(methanesulfonyl)amino]-1-methyl-1H-indazol-7-yl}-7-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-4-oxo-3,4-dihydroquinazolin-2-yl}-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-3-(difluoromethyl)-5,5-difluoro-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Nolte, R.T. | | Deposit date: | 2022-12-16 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Potent Long-Acting Inhibitors Targeting the HIV-1 Capsid Based on a Versatile Quinazolin-4-one Scaffold.
J.Med.Chem., 66, 2023
|
|
7NVK
 
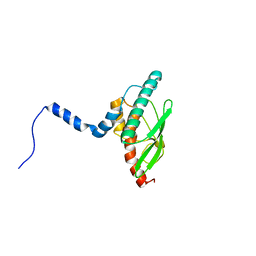 | | Crystal structure of UBA5 fragment fused to the N-terminus of UFC1 | | Descriptor: | Ubiquitin-like modifier-activating enzyme 5,Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
8G0E
 
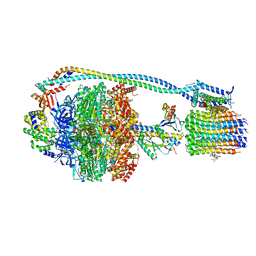 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0C
 
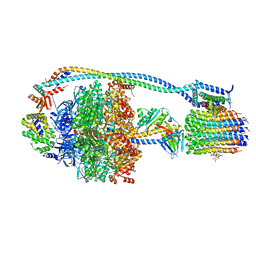 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
6F88
 
 | |
6F9C
 
 | |
8RZB
 
 | | IL-1beta in complex with covalent DEL hit | | Descriptor: | 8-[4-methyl-3-(trifluoromethyl)phenyl]-2-[[(7S)-7-(2-morpholin-4-ylethylcarbamoyl)-4-(phenylsulfonyl)-1,4-diazepan-1-yl]carbonyl]imidazo[1,2-a]pyridine-6-carboxylic acid, Interleukin-1 beta | | Authors: | Rondeau, J.-M, Lehmann, S. | | Deposit date: | 2024-02-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Ligandability Assessment of IL-1 beta by Integrated Hit Identification Approaches.
J.Med.Chem., 67, 2024
|
|
8RS9
 
 | | p97 (VCP) double mutant - F266A F539A | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | p97 (VCP) double mutant - F266A F539A
To Be Published
|
|
8G0D
 
 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8FRC
 
 | |
4LCB
 
 | | Structure of Vps4 homolog from Acidianus hospitalis | | Descriptor: | CHLORIDE ION, Cell division protein CdvC, Vps4 | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-21 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
6EW9
 
 | | CRYSTAL STRUCTURE OF DEGS STRESS SENSOR PROTEASE IN COMPLEX WITH ACTIVATING DNRLGLVYQF PEPTIDE | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DNRLGLVYQF PEPTIDE, ... | | Authors: | Vetter, I.R, Porfetye, A.T, Stege, P. | | Deposit date: | 2017-11-03 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Noncatalytic Lysine Residues from Allosteric Circuits via Covalent Probes.
ACS Chem. Biol., 13, 2018
|
|
8FFD
 
 | | Crystal structure of manganeese bound Dps protein (PA0962) from Pseudomonas aeruginosa (cubic form) | | Descriptor: | L(+)-TARTARIC ACID, MANGANESE (II) ION, Probable dna-binding stress protein | | Authors: | Lovell, S, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8G0A
 
 | |
4KVD
 
 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (4aS,7S)-1,4a-dimethyl-7-(prop-1-en-2-yl)decahydroquinolin-1-ium | | Descriptor: | (1R,4aS,7S,8aR)-1,4a-dimethyl-7-(prop-1-en-2-yl)decahydroquinolinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Faraldos, J.A, Al-lami, N, Janvier, M, D'Antonio, E.L, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4KVX
 
 | | Crystal structure of Naa10 (Ard1) bound to AcCoA | | Descriptor: | ACETYL COENZYME *A, N-terminal acetyltransferase A complex catalytic subunit ard1 | | Authors: | Liszczak, G.P, Marmorstein, R.Q. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for N-terminal acetylation by the heterodimeric NatA complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8FUB
 
 | | Crystal structure of human Importin alpha 3 in complex with Hendra virus matrix protein NLS1 | | Descriptor: | Importin subunit alpha-3, Matrix protein | | Authors: | Donnelly, C.M, Basler, C.F, Scott, C, Forwood, J.K. | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Henipavirus Matrix Protein Employs a Non-Classical Nuclear Localization Signal Binding Mechanism.
Viruses, 15, 2023
|
|
