6T8M
 
 | | Prolyl Hydroxylase (PHD) involved in hypoxia sensing by Dictyostelium discoideum | | Descriptor: | CHLORIDE ION, GLYCEROL, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, McDonough, M.A, Liu, T, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2019-10-24 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and biophysical analyses of hypoxia sensing prolyl hydroxylases from Dictyostelium discoideum and Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
9GM3
 
 | | Crystal structure of the complex formed between the radical SAM protein ChlB and the leader region of its precursor substrate ChlA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ChlA, ChlB radical SAM domain, ... | | Authors: | de la Mora, E, Ruel, J, Usclat, A, Martin, L, Amara, P, Morinaka, B, Nicolet, Y. | | Deposit date: | 2024-08-28 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Peptide Recognition and Mechanism of the Radical S -Adenosyl-l-methionine Multiple Cyclophane Synthase ChlB.
J.Am.Chem.Soc., 147, 2025
|
|
8A9P
 
 | | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment | | Descriptor: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment
To Be Published
|
|
9M3M
 
 | | Structure of FSP1 in complex with FSEN1 | | Descriptor: | 3-(4-bromophenyl)-6-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-[1,3]thiazolo[2,3-c][1,2,4]triazole, 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, ... | | Authors: | Zhang, S.T, Jia, D. | | Deposit date: | 2025-03-03 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cocrystal structure reveals the mechanism of FSP1 inhibition by FSEN1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
3HSO
 
 | | Ternary structure of neuronal nitric oxide synthase with NHA and NO bound(1) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-HYDROXY-L-ARGININE, ... | | Authors: | Doukov, T, Li, H, Soltis, M, Poulos, T.L. | | Deposit date: | 2009-06-10 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Single crystal structural and absorption spectral characterizations of nitric oxide synthase complexed with N(omega)-hydroxy-L-arginine and diatomic ligands.
Biochemistry, 48, 2009
|
|
6E87
 
 | |
8RR0
 
 | | CryoEM structure of Molybdenum bispyranopterin guanine dinucleotide formate dehydrogenases ForCE1 from Bacillus subtilis | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, HEPTANOIC ACID, ... | | Authors: | Cherrier, M.V, Arnoux, P, Martin, L, Nicolet, Y, Schoehn, G, Legrand, P, Broc, M, Seduk, F, Brasseur, G, Arias-Cartin, R, Magalon, A, Walburger, A, Uzel, A, Guigliarelli, B, Grimaldi, S, Pierrel, F, Mate, M. | | Deposit date: | 2024-01-22 | | Release date: | 2025-08-06 | | Last modified: | 2025-09-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | A scaffold for quinone channeling between membrane and soluble bacterial oxidoreductases.
Nat.Struct.Mol.Biol., 2025
|
|
3HSN
 
 | | Ternary structure of neuronal nitric oxide synthase with NHA and CO bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CARBON MONOXIDE, ... | | Authors: | Doukov, T, Li, H, Soltis, M, Poulos, T.L. | | Deposit date: | 2009-06-10 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Single crystal structural and absorption spectral characterizations of nitric oxide synthase complexed with N(omega)-hydroxy-L-arginine and diatomic ligands.
Biochemistry, 48, 2009
|
|
7TS8
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 4-chloro-6-((5-(2-(dimethylamino)ethyl)-2,3-difluorophenyl)ethynyl)pyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[3-(dimethylamino)propyl]-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Aminopyridines with a shortened amino sidechain as potent, selective, and highly permeable human neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem., 69, 2022
|
|
7V0B
 
 | |
8PL1
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni fragment screen hit 2. | | Descriptor: | 5-(methoxymethyl)-1,3,4-thiadiazol-2-amine, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-27 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
8PLR
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni fragment screen hit 28. | | Descriptor: | (2R)-1-{[(5-methylthiophen-2-yl)methyl]amino}propan-2-ol, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-27 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
8BGW
 
 | | CryoEM structure of quinol-dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans at 2.2 A resolution | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, FE (III) ION, ... | | Authors: | Flynn, A, Antonyuk, S.V, Eady, R.R, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2022-10-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | A 2.2 angstrom cryoEM structure of a quinol-dependent NO Reductase shows close similarity to respiratory oxidases.
Nat Commun, 14, 2023
|
|
6Y77
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H326A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINUCLEAR COPPER ION, FORMIC ACID, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
9C0Q
 
 | |
9B17
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PAN1 inhibitor | | Descriptor: | (5P)-5-(1H-indol-3-yl)-1-[2-(piperazin-1-yl)ethyl]-1H-1,2,3-benzotriazole, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Geeraerts, Z, Yeh, S.-R. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insights into Protein-Inhibitor Interactions in Human Tryptophan Dioxygenase.
J.Med.Chem., 67, 2024
|
|
9B1Q
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PYN1 inhibitor | | Descriptor: | (5P)-1-[(imidazolidin-1-yl)methyl]-5-(1H-indol-3-yl)-1H-1,2,3-benzotriazole, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Geeraerts, Z, Yeh, S.-R. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.624 Å) | | Cite: | Structural Insights into Protein-Inhibitor Interactions in Human Tryptophan Dioxygenase.
J.Med.Chem., 67, 2024
|
|
9AT2
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PAN3 inhibitor | | Descriptor: | (6M)-6-(1H-indol-3-yl)-1-[2-(piperazin-1-yl)ethyl]-1H-1,2,3-benzotriazole, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Geeraerts, Z, Yeh, S.-R. | | Deposit date: | 2024-02-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into Protein-Inhibitor Interactions in Human Tryptophan Dioxygenase.
J.Med.Chem., 67, 2024
|
|
6SYM
 
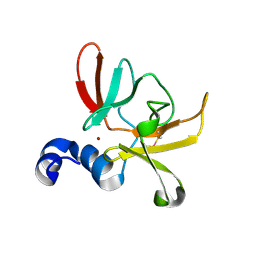 | |
8CJY
 
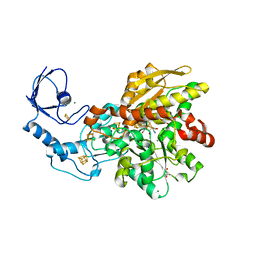 | | [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant S357T | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Brocks, C, Duan, J, Hofmann, E, Happe, T. | | Deposit date: | 2023-02-13 | | Release date: | 2023-10-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Dynamic Water Channel Affects O 2 Stability in [FeFe]-Hydrogenases.
Chemsuschem, 17, 2024
|
|
8CVD
 
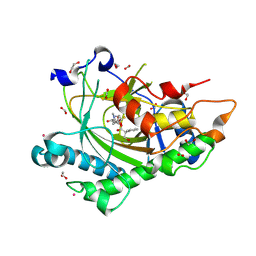 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and scopolamine | | Descriptor: | (1R,2R,4S,5S,7s)-9-methyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-7-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVA
 
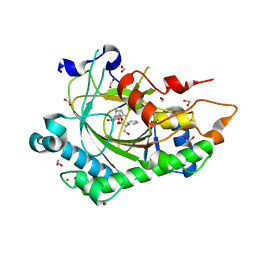 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8VHL
 
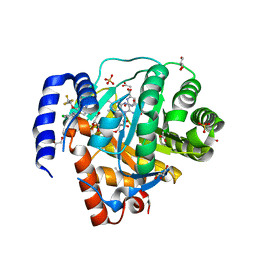 | | Structure of DHODH in Complex with Ligand 17 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8RFN
 
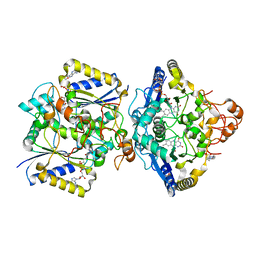 | | Human NOQ1 enzyme in its holo form by serial crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Medina, M, Boneta, S, Pey, A.L. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural dynamics and functional cooperativity of human NQO1 by ambient temperature serial crystallography and simulations.
Protein Sci., 33, 2024
|
|
5MOG
 
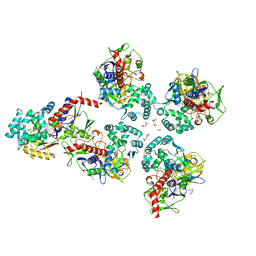 | | Oryza sativa phytoene desaturase inhibited by norflurazon | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brausemann, A, Gemmecker, S, Koschmieder, J, Beyer, P, Einsle, O. | | Deposit date: | 2016-12-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure of Phytoene Desaturase Provides Insights into Herbicide Binding and Reaction Mechanisms Involved in Carotene Desaturation.
Structure, 25, 2017
|
|
