6C61
 
 | | MHC-independent T-cell receptor B12A | | Descriptor: | T-cell receptor alpha chain, T-cell receptor beta chain | | Authors: | Lu, J, Sun, P. | | Deposit date: | 2018-01-17 | | Release date: | 2019-01-30 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of MHC-Independent TCRs and Their Recognition of Native Antigen CD155.
J Immunol., 204, 2020
|
|
6C68
 
 | | MHC-independent t cell receptor A11 | | Descriptor: | T-cell receptor alpha chain, T-cell receptor beta chain | | Authors: | Lu, J, Van Laethem, F, Saba, I, Chu, J, Bhattacharya, A, Love, N.C, Tikhonova, A, Radaev, S, Sun, X, Ko, A, Arnon, T, Shifrut, E, Friedman, N, Weng, N, Singer, A, Sun, P.D. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of MHC-Independent TCRs and Their Recognition of Native Antigen CD155.
J Immunol., 204, 2020
|
|
8U5L
 
 | |
4LP4
 
 | |
4ZHC
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | ACETATE ION, N-{2-[bis(2-{[(2,3-dihydroxyphenyl)carbonyl]amino}ethyl)amino]ethyl}-1-hydroxy-6-oxo-1,6-dihydropyridine-2-carboxamide, Neutrophil gelatinase-associated lipocalin, ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4LP5
 
 | |
4ZHF
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | CURIUM ION, GLYCEROL, N,N'-butane-1,4-diylbis[1-hydroxy-N-(3-{[(1-hydroxy-6-oxo-1,6-dihydropyridin-2-yl)carbonyl]amino}propyl)-6-oxo-1,6-dihydropyridine-2-carboxamide], ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7R20
 
 | | Anti-Arc nanobody E5 | | Descriptor: | Anti-Arc nanobody E5, GLYCEROL, SULFATE ION | | Authors: | Markusson, S, Kursula, P. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-affinity anti-Arc nanobodies provide tools for structural and functional studies.
Plos One, 17, 2022
|
|
7R1Z
 
 | |
8DCI
 
 | |
2OP4
 
 | | Crystal Structure of Quorum-Quenching Antibody 1G9 | | Descriptor: | 1,2-ETHANEDIOL, Murine Antibody Fab RS2-1G9 IGG1 Heavy Chain, Murine Antibody Fab RS2-1G9 Lambda Light Chain | | Authors: | Kirchdoerfer, R.N, Debler, E.W, Wilson, I.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of a Quorum-quenching Antibody.
J.Mol.Biol., 368, 2007
|
|
4GPB
 
 | |
8BQP
 
 | | Hen Egg-White Lysozyme (HEWL) complexed with methyl-functionalised Anderson-Evans polyoxometalate | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lentink, S, Salazar Marcano, D.E, Moussawi, M.A, Vandebroek, L, Van Meervelt, L, Parac-Vogt, T.N. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Fine-tuning non-covalent interactions between hybrid metal-oxo clusters and proteins.
Faraday Disc.Chem.Soc, 244, 2023
|
|
8BQQ
 
 | | Hen Egg-White Lysozyme (HEWL) complexed with amine-functionalised Anderson-Evans polyoxometalate | | Descriptor: | CHLORIDE ION, Lysozyme C, Mn-Mo(6)-N(2)-O(24)-C(8) cluster | | Authors: | Lentink, S, Salazar Marcano, D.E, Moussawi, M.A, Vandebroek, L, Van Meervelt, L, Parac-Vogt, T.N. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Fine-tuning non-covalent interactions between hybrid metal-oxo clusters and proteins.
Faraday Disc.Chem.Soc, 244, 2023
|
|
8BQR
 
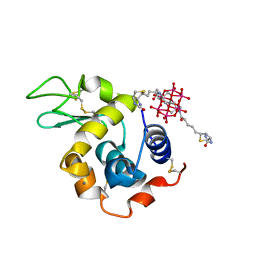 | | Hen Egg-White Lysozyme (HEWL) complexed with biotin-functionalised Anderson-Evans polyoxometalate | | Descriptor: | Anderson-Evans polyoxometalate (biotin-functionalised), CALCIUM ION, Lysozyme C | | Authors: | Lentink, S, Salazar Marcano, D.E, Moussawi, M.A, Vandebroek, L, Van Meervelt, L, Parac-Vogt, T.N. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Fine-tuning non-covalent interactions between hybrid metal-oxo clusters and proteins.
Faraday Disc.Chem.Soc, 244, 2023
|
|
8BQT
 
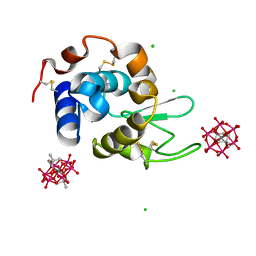 | | Hen Egg-White Lysozyme (HEWL) complexed with two methyl-functionalised Anderson-Evans polyoxometalates | | Descriptor: | CHLORIDE ION, Lysozyme C, Mn-Mo(6)-O(24)-C(10) cluster | | Authors: | Lentink, S, Salazar Marcano, D.E, Moussawi, M.A, Vandebroek, L, Van Meervelt, L, Parac-Vogt, T.N. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Fine-tuning non-covalent interactions between hybrid metal-oxo clusters and proteins.
Faraday Disc.Chem.Soc, 244, 2023
|
|
7B9H
 
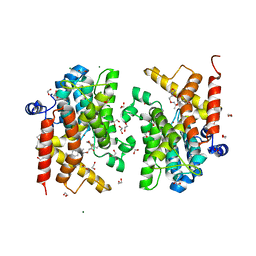 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-42a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-1-(3-cyclopentyloxy-4-methoxy-phenyl)ethylideneamino]oxy-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ... | | Authors: | Torretta, A, Abbate, S, Parisini, E. | | Deposit date: | 2020-12-14 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, biological evaluation and structural characterization of novel GEBR library PDE4D inhibitors.
Eur.J.Med.Chem., 223, 2021
|
|
5KHA
 
 | |
3UON
 
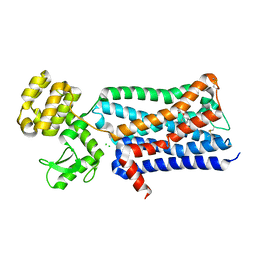 | | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, CHLORIDE ION, Human M2 muscarinic acetylcholine, ... | | Authors: | Haga, K, Kruse, A.C, Asada, H, Yurugi-Kobayashi, T, Shiroishi, M, Zhang, C, Weis, W.I, Okada, T, Kobilka, B.K, Haga, T, Kobayashi, T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist.
Nature, 482, 2012
|
|
7S7K
 
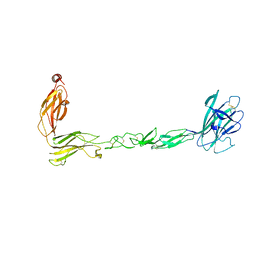 | | Crystal structure of the EphB2 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 2, ... | | Authors: | Xu, Y, Xu, K, Nikolov, D.B. | | Deposit date: | 2021-09-16 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The Ephb2 Receptor Uses Homotypic, Head-to-Tail Interactions within Its Ectodomain as an Autoinhibitory Control Mechanism.
Int J Mol Sci, 22, 2021
|
|
8UF3
 
 | |
8SG1
 
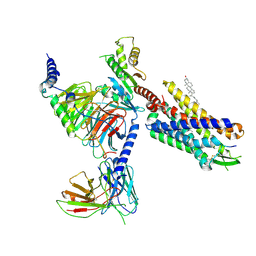 | | Cryo-EM structure of CMKLR1 signaling complex | | Descriptor: | CHOLESTEROL, Chemerin 9, Chemerin-like receptor 1, ... | | Authors: | Zhang, X, Zhang, C. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of G protein-Coupled receptor CMKLR1 activation and signaling induced by a chemerin-derived agonist.
Plos Biol., 21, 2023
|
|
5L9T
 
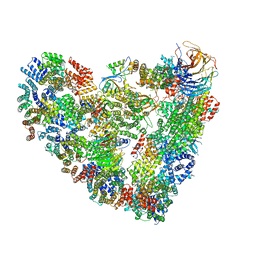 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
8OO6
 
 | | Pol I bound to extended and displaced DNA section - closed conformation | | Descriptor: | DNA polymerase I, Displaced primer, Extending Primer, ... | | Authors: | Botto, M, Borsellini, A, Lamers, M.H. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OOY
 
 | | Pol I bound to extended and displaced DNA section - open conformation | | Descriptor: | DNA polymerase I, Displacing Primer, Extending Primer, ... | | Authors: | Botto, M, Borsellini, A, Lamers, M.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
