8SC2
 
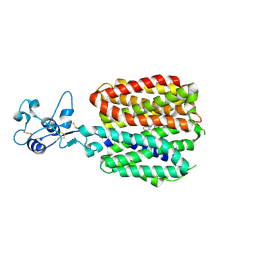 | |
8SC6
 
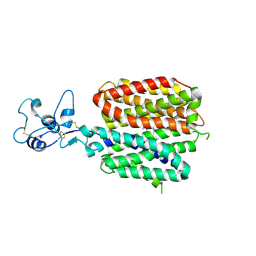 | | Human OCT1 bound to thiamine in inward-open conformation | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Solute carrier family 22 member 1 | | Authors: | Zeng, Y.C, Sobti, M, Stewart, A.G. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis of promiscuous substrate transport by Organic Cation Transporter 1.
Nat Commun, 14, 2023
|
|
424D
 
 | | 5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3' | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3') | | Authors: | Rozenberg, H, Rabinovich, D, Frolow, F, Hegde, R.S, Shakked, Z. | | Deposit date: | 1998-09-14 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6CMW
 
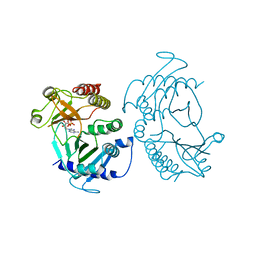 | | Crystal structure of zebrafish Phosphatidylinositol-4-phosphate 5- kinase alpha isoform with bound ATP/Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol-4-phosphate 5-kinase, ... | | Authors: | Zeng, X, Sui, D, Hu, J. | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into lethal contractural syndrome type 3 (LCCS3) caused by a missense mutation of PIP5K gamma.
Biochem. J., 475, 2018
|
|
6NWU
 
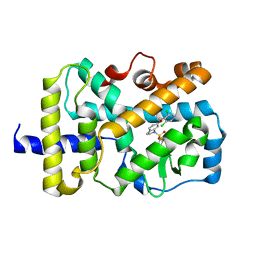 | | RORgamma Ligand Binding Domain | | Descriptor: | 6-[(3,5-dichloropyridin-4-yl)methoxy]-1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indole, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWT
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
425D
 
 | | 5'-D(*AP*CP*CP*GP*GP*TP*AP*CP*CP*GP*GP*T)-3' | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*TP*AP*CP*CP*GP*GP*T)-3') | | Authors: | Rozenberg, H, Rabinovich, D, Frolow, F, Hegde, R.S, Shakked, Z. | | Deposit date: | 1998-09-14 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6CN3
 
 | |
1LOU
 
 | | RIBOSOMAL PROTEIN S6 | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Otzen, D.E, Kristensen, O, Proctor, M, Oliveberg, M. | | Deposit date: | 1998-11-25 | | Release date: | 1998-11-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural changes in the transition state of protein folding: alternative interpretations of curved chevron plots.
Biochemistry, 38, 1999
|
|
1D2N
 
 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
1LXG
 
 | |
1LXH
 
 | |
8SWJ
 
 | | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531 | | Descriptor: | (3S)-N-(4'-carbamoyl[1,1'-biphenyl]-3-yl)-1-[4-(4-methylpiperazin-1-yl)pyridine-2-carbonyl]piperidine-3-carboxamide, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Shell, D.J, Foley, C, Pearce, K.H, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531
To be published
|
|
2MFQ
 
 | |
2YJ0
 
 | | X-ray structure of chemically engineered Mycobacterium tuberculosis Dodecin | | Descriptor: | CHLORIDE ION, COENZYME A, DODECIN, ... | | Authors: | Vinzenz, X, Grosse, W, Linne, U, Meissner, B, Essen, L.-O. | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical Engineering of Mycobacterium Tuberculosis Dodecin Hybrids.
Chem.Commun.(Camb.), 47, 2011
|
|
8D41
 
 | | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254 | | Descriptor: | (1R,2R,3S,4S)-3-[4-(4-fluorophenyl)piperazine-1-carbonyl]bicyclo[2.2.1]heptane-2-carboxylic acid, Coatomer subunit beta', GLYCEROL, ... | | Authors: | Zeng, H, Saraon, P, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254
To Be Published
|
|
2YCE
 
 | | Structure of an Archaeal fructose-1,6-bisphosphate aldolase with the catalytic Lys covalently bound to the carbinolamine intermediate of the substrate. | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS 1 | | Authors: | Lorentzen, E, Siebers, B, Hensel, R, Pohl, E. | | Deposit date: | 2011-03-14 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of the Schiff Base Forming Fructose-1,6-Bisphosphate Aldolase: Structural Analysis of Reaction Intermediates.
Biochemistry, 44, 2005
|
|
8HVK
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF07321332
To Be Published
|
|
8HQF
 
 | |
8D30
 
 | | Crystal structure of the human COPB2 WD-domains | | Descriptor: | 1,2-ETHANEDIOL, Coatomer subunit beta' | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-31 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human COPB2 WD-domains
To Be Published
|
|
1YB5
 
 | | Crystal structure of human Zeta-Crystallin with bound NADP | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Debreczeni, J, Berridge, G, Kavanagh, K, Colbrook, S, Bray, J, Williams, L, Oppermann, U, Sundstrom, M, Arrowsmith, C, Edwards, A, Gileadi, O, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-20 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of human Zeta-Crystallin at 1.85A
To be Published
|
|
1W8G
 
 | | CRYSTAL STRUCTURE OF E. COLI K-12 YGGS | | Descriptor: | HYPOTHETICAL UPF0001 PROTEIN YGGS, ISOCITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sulzenbacher, G, Gruez, A, Spinelli, S, Roig-Zamboni, V, Pagot, F, Bignon, C, Vincentelli, R, Cambillau, C. | | Deposit date: | 2004-09-21 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of E. Coli K-12 Yggs
To be Published
|
|
1XBP
 
 | | Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with Tiamulin | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Schluenzen, F, Pyetan, E, Fucini, P, Yonath, A, Harms, J.M. | | Deposit date: | 2004-08-31 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin.
Mol.Microbiol., 54, 2004
|
|
2BXJ
 
 | | Double Mutant of the Ribosomal Protein S6 | | Descriptor: | 30S RIBOSOMAL PROTEIN S6 | | Authors: | Otzen, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antagonism, Non-Native Interactions and Non-Two-State Folding in S6 Revealed by Double-Mutant Cycle Analysis.
Protein Eng.Des.Sel., 18, 2005
|
|
8HZR
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF07321332
To Be Published
|
|
