8RDV
 
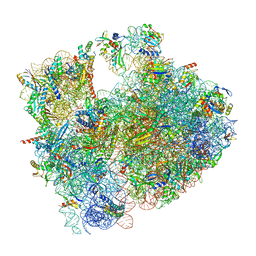 | | Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2). | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Helena-Bueno, K, Rybak, M.Y, Gagnon, M.G, Hill, C.H, Melnikov, S.V. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8V9B
 
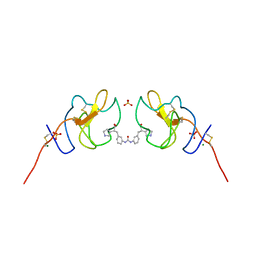 | | Lipoprotein(a) Kringle IV domain 7 - Lp(a) KIV7 in complex with LY3441732 | | Descriptor: | (2S,2'S)-3,3'-[carbonylbis(azanediyl-3,1-phenylene)]bis{2-[(3R)-pyrrolidin-1-ium-3-yl]propanoate}, Apolipoprotein(a), MAGNESIUM ION, ... | | Authors: | Hendle, J, Weichert, K, Sauder, J.M. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Discovery of potent small-molecule inhibitors of lipoprotein(a) formation.
Nature, 629, 2024
|
|
8RD8
 
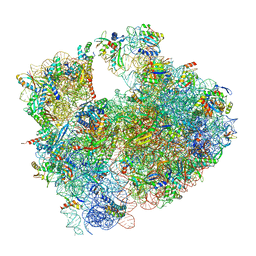 | | Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S30, ... | | Authors: | Helena-Bueno, K, Rybak, M.Y, Gagnon, M.G, Hill, C.H, Melnikov, S.V. | | Deposit date: | 2023-12-07 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8XBE
 
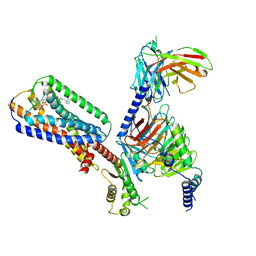 | | Human GPR34 -Gi complex bound to S3E-LysoPS | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[(~{Z})-octadec-9-enoyl]oxy-propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBG
 
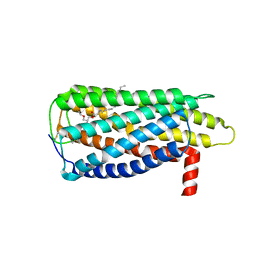 | |
8V8U
 
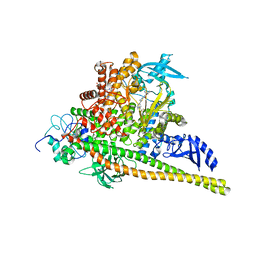 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket near H1047R (compound 12). | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, (3S)-9-[(1R)-1-(2-carboxyanilino)ethyl]-3-cyano-7-methyl-4-oxo-2-(piperidin-1-yl)-3,4-dihydropyrido[1,2-a]pyrimidin-5-ium, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8V
 
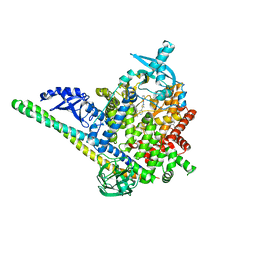 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket near H1047R (compound 7). | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, 2-[[(1~{R})-1-(7-methyl-4-oxidanylidene-2-piperidin-1-yl-3~{H}-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8Z
 
 | |
8XBI
 
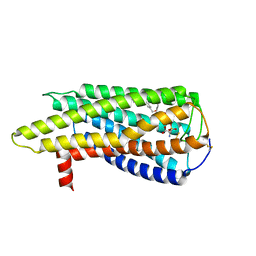 | | Human GPR34 -Gi complex bound to M1, receptor focused | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Probable G-protein coupled receptor 34 | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBH
 
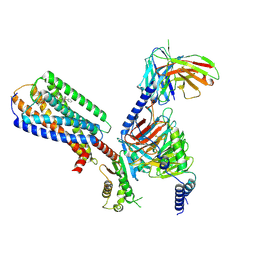 | | Human GPR34 -Gi complex bound to M1 | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8V8I
 
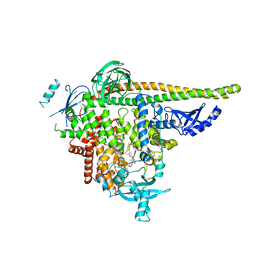 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket (compound 5). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, CHLORIDE ION, N-{(3S)-3-(2-methylphenyl)-6-[(oxetan-3-yl)amino]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-1-benzothiophene-3-carboxamide, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8XAT
 
 | |
8V8H
 
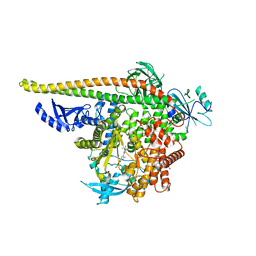 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket near H1047R (compound 4). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, 2-({(1R)-1-[2-(4,4-dimethylpiperidin-1-yl)-3,6-dimethyl-4-oxo-4H-1-benzopyran-8-yl]ethyl}amino)benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8J
 
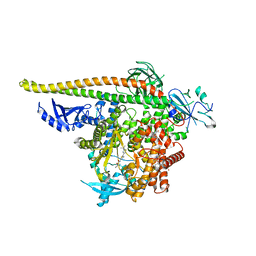 | | PI3Ka H1047R co-crystal structure with inhibitors in two cryptic pockets (compounds 4 and 5). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, 2-({(1R)-1-[2-(4,4-dimethylpiperidin-1-yl)-3,6-dimethyl-4-oxo-4H-1-benzopyran-8-yl]ethyl}amino)benzoic acid, N-{(3S)-3-(2-methylphenyl)-6-[(oxetan-3-yl)amino]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-1-benzothiophene-3-carboxamide, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8RC0
 
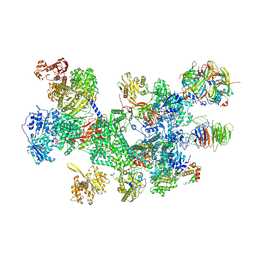 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8RC1
 
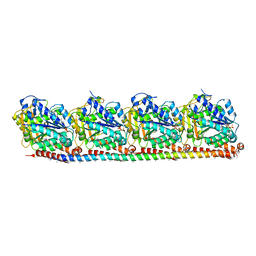 | |
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V83
 
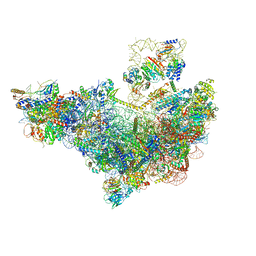 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V6U
 
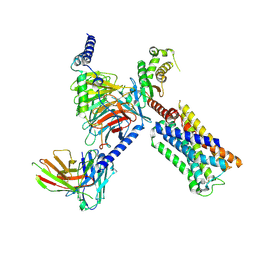 | | 5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking | | Descriptor: | 4-{(3R)-1-[(1R)-1-(pyrimidin-2-yl)ethyl]piperidin-3-yl}phenol, 5-hydroxytryptamine receptor 2A, G protein alpha-subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Gumpper, R.H, Wang, L, Kapolka, N, Skiniotis, G, Roth, B.L. | | Deposit date: | 2023-12-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
8V6N
 
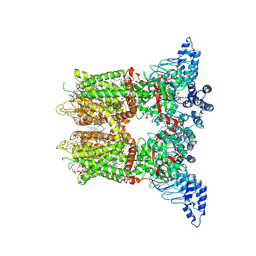 | | Open-state cryo-EM structure of human TRPV3 in presence of 2-APB in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-aminoethyl diphenylborinate, ARACHIDONIC ACID, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
8V6M
 
 | | Inactivated-state cryo-EM structure of human TRPV3 in presence of tetrahydrocannabivarin (THCV) in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Tetrahydrocannabivarin, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
8V6L
 
 | | Open-state cryo-EM structure of human TRPV3 in presence of tetrahydrocannabivarin (THCV) in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Tetrahydrocannabivarin, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
8V6K
 
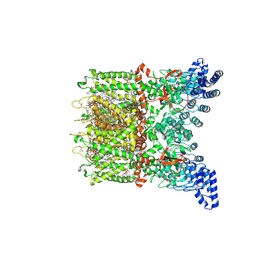 | | Apo-state cryo-EM structure of human TRPV3 in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
8V6O
 
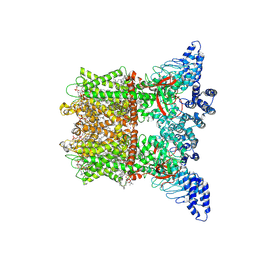 | | Inactivated-state cryo-EM structure of human TRPV3 in presence of 2-APB in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-aminoethyl diphenylborinate, SODIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
