3SC7
 
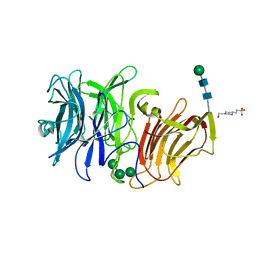 | | First crystal structure of an endo-inulinase, from Aspergillus ficuum: structural analysis and comparison with other GH32 enzymes. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Inulinase, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, ... | | Authors: | Housen, I, Pouyez, J, Roussel, G, Mayard, A, Vandamme, A.M, Wouters, J, Michaux, C. | | Deposit date: | 2011-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: Discovery of an extra-pocket in the catalytic domain responsible for its endo-activity.
Biochimie, 94, 2012
|
|
4I7I
 
 | |
4I0Y
 
 | |
1ZWA
 
 | |
1ZWC
 
 | |
1D3I
 
 | |
2OB1
 
 | | ppm1 with 1,8-ANS | | Descriptor: | Leucine carboxyl methyltransferase 1, PHOSPHATE ION | | Authors: | Groves, M.R. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A method for the general identification of protein crystals in crystallization experiments using a noncovalent fluorescent dye.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1GA2
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM COMPLEXED WITH CELLOBIOSE | | Descriptor: | ACETIC ACID, CALCIUM ION, ENDOGLUCANASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-29 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
2O62
 
 | |
4R2G
 
 | | Crystal Structure of PGT124 Fab bound to HIV-1 JRCSF gp120 core and to CD4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Garces, F, Wilson, I.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
2B1I
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, [3,4-DIHYDROXY-5R-(2,2,4-TRIOXO-1,2R,3S,4R-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-7-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
3RAI
 
 | | CDK2 in complex with inhibitor KVR-1-160 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[3-(morpholin-4-yl)propyl]amino}-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R1Y
 
 | | CDK2 in complex with inhibitor KVR-1-134 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-11 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
2NAV
 
 | | NMR solution structure of Ex-4[1-16]/pl14a | | Descriptor: | Exendin-4, Alpha/kappa-conotoxin pl14a chimera | | Authors: | Schroeder, C.I, Swedberg, J.E, Craik, D.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Truncated Glucagon-like Peptide-1 and Exendin-4 alpha-Conotoxin pl14a Peptide Chimeras Maintain Potency and alpha-Helicity and Reveal Interactions Vital for cAMP Signaling in Vitro.
J.Biol.Chem., 291, 2016
|
|
3NXD
 
 | | JC polyomavirus VP1 in complex with LSTc | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1, ... | | Authors: | Neu, U, Stroeh, L.J, Stehle, T. | | Deposit date: | 2010-07-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif.
Cell Host Microbe, 8, 2010
|
|
5TZS
 
 | | Architecture of the yeast small subunit processome | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 3' domain-associated, ... | | Authors: | Chaker-Margot, M, Barandun, J, Hunziker, M, Klinge, S. | | Deposit date: | 2016-11-22 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Architecture of the yeast small subunit processome.
Science, 355, 2017
|
|
3SAL
 
 | | Crystal Structure of Influenza A Virus Neuraminidase N5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, M.Y, Qi, J.X, Liu, Y, Vavricka, C.J, Wu, Y, Li, Q, Gao, G.F. | | Deposit date: | 2011-06-03 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influenza a virus n5 neuraminidase has an extended 150-cavity
J.Virol., 85, 2011
|
|
2NW6
 
 | | Burkholderia cepacia lipase complexed with S-inhibitor | | Descriptor: | (1S)-1-(PHENOXYMETHYL)PROPYL METHYLPHOSPHONOCHLORIDOATE, CALCIUM ION, Lipase, ... | | Authors: | Luic, M, Stefanic, Z. | | Deposit date: | 2006-11-14 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined X-ray diffraction and QM/MM study of the Burkholderia cepacia lipase-catalyzed secondary alcohol esterification
J.Phys.Chem.B, 112, 2008
|
|
2B1G
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
4DOE
 
 | | The liganded structure of Cbescii CelA GH9 module | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1,4-beta-glucanase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-02-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA.
Science, 342, 2013
|
|
4JPM
 
 | |
2OJN
 
 | |
1Y9M
 
 | | Crystal structure of exo-inulinase from Aspergillus awamori in spacegroup P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nagem, R.A.P, Rojas, A.L, Golubev, A.M, Korneeva, O.S, Eneyskaya, E.V, Kulminskaya, A.A, Neustroev, K.N, Polikarpov, I. | | Deposit date: | 2004-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition
J.Mol.Biol., 344, 2004
|
|
1ULV
 
 | | Crystal Structure of Glucodextranase Complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
1SMB
 
 | | Crystal Structure of Golgi-Associated PR-1 protein | | Descriptor: | 17kD fetal brain protein | | Authors: | Serrano, R.L, Kuhn, A, Hendricks, A, Helms, J.B, Sinning, I, Groves, M.R. | | Deposit date: | 2004-03-08 | | Release date: | 2004-09-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of the human Golgi-associated plant pathogenesis related protein GAPR-1 implicates dimerization as a regulatory mechanism
J.Mol.Biol., 339, 2004
|
|
