8C5D
 
 | | Glutathione transferase P1-1 from Mus musculus | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2023-01-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Inhibition Analysis and High-Resolution Crystal Structure of Mus musculus Glutathione Transferase P1-1.
Biomolecules, 13, 2023
|
|
5GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-11 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
5J41
 
 | | Glutathione S-transferase bound with hydrolyzed Piperlongumine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(3,4,5-trimethoxyphenyl)propanoic acid, GLUTATHIONE, ... | | Authors: | Harshbarger, W, Gondi, S, Ficarro, S, Hunter, J, Udayakumar, D, Gurbani, D, Marto, J, Westover, K. | | Deposit date: | 2016-03-31 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.19035351 Å) | | Cite: | Structural and Biochemical Analyses Reveal the Mechanism of Glutathione S-Transferase Pi 1 Inhibition by the Anti-cancer Compound Piperlongumine.
J. Biol. Chem., 292, 2017
|
|
8WMH
 
 | | Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMM
 
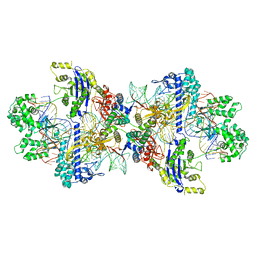 | | Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary) | | Descriptor: | MAGNESIUM ION, NTS, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMN
 
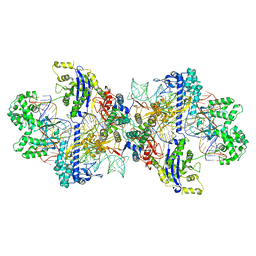 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary) | | Descriptor: | DNA (62-MER), MAGNESIUM ION, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WR4
 
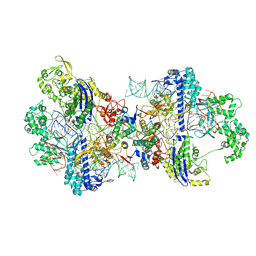 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex) | | Descriptor: | CbCas9 effector-1, DNA (62-MER), MAGNESIUM ION, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
6ZMN
 
 | |
2PGT
 
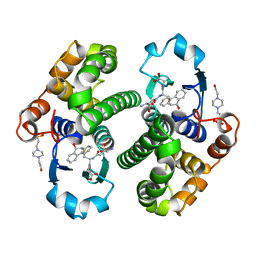 | | CRYSTAL STRUCTURE OF HUMAN GLUTATHIONE S-TRANSFERASE P1-1[V104] COMPLEXED WITH (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, ... | | Authors: | Ji, X. | | Deposit date: | 1997-02-17 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate-binding site and location of a potential non-substrate-binding site in a class pi glutathione S-transferase.
Biochemistry, 36, 1997
|
|
6AP9
 
 | |
18GS
 
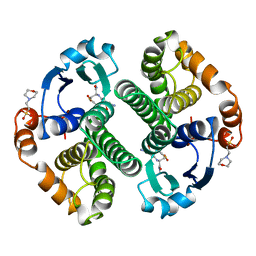 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH 1-(S-GLUTATHIONYL)-2,4-DINITROBENZENE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-12-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
10GS
 
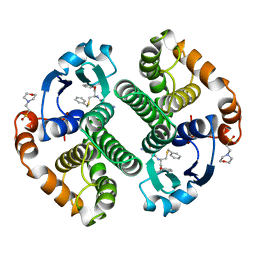 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH TER117 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, L-gamma-glutamyl-S-benzyl-N-[(S)-carboxy(phenyl)methyl]-L-cysteinamide | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
11GS
 
 | | Glutathione s-transferase complexed with ethacrynic acid-glutathione conjugate (form ii) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Mazzetti, A.P, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The glutathione conjugate of ethacrynic acid can bind to human pi class glutathione transferase P1-1 in two different modes.
FEBS Lett., 419, 1997
|
|
16GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 APO FORM 3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence for an induced-fit mechanism operating in pi class glutathione transferases.
Biochemistry, 37, 1998
|
|
19GS
 
 | | Glutathione s-transferase p1-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,3'-(4,5,6,7-TETRABROMO-3-OXO-1(3H)-ISOBENZOFURANYLIDENE)BIS [6-HYDROXYBENZENESULFONIC ACID]ANION, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-12-14 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
12GS
 
 | | GLUTATHIONE S-TRANSFERASE COMPLEXED WITH S-NONYL-GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-nonyl-L-cysteinylglycine | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-11-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Ligandin (Non-Substrate) Binding Site of Human Pi Class Glutathione Transferase is Located in the Electrophile Binding Site (H-Site).
J.Mol.Biol., 291, 1999
|
|
13GS
 
 | | GLUTATHIONE S-TRANSFERASE COMPLEXED WITH SULFASALAZINE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-11-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Ligandin (Non-Substrate) Binding Site of Human Pi Class Glutathione Transferase is Located in the Electrophile Binding Site (H-Site).
J.Mol.Biol., 291, 1999
|
|
1AQV
 
 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH P-BROMOBENZYLGLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, N-[(4S)-4-ammonio-4-carboxybutanoyl]-S-(4-bromobenzyl)-L-cysteinylglycine | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-01 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
1AQW
 
 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
14GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 APO FORM 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for an induced-fit mechanism operating in pi class glutathione transferases.
Biochemistry, 37, 1998
|
|
17GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-12-07 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glutathione S-transferase P1-1
To be published
|
|
1AQX
 
 | | GLUTATHIONE S-TRANSFERASE IN COMPLEX WITH MEISENHEIMER COMPLEX | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Prade, L, Huber, R, Manoharan, T.H, Fahl, W.E, Reuter, W. | | Deposit date: | 1997-08-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor.
Structure, 5, 1997
|
|
1BAY
 
 | | GLUTATHIONE S-TRANSFERASE YFYF CYS 47-CARBOXYMETHYLATED CLASS PI, FREE ENZYME | | Descriptor: | GLUTATHIONE S-TRANSFERASE CLASS PI | | Authors: | Vega, M.C, Coll, M. | | Deposit date: | 1996-11-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of Cys-47-modified mouse liver glutathione S-transferase P1-1. Carboxymethylation dramatically decreases the affinity for glutathione and is associated with a loss of electron density in the alphaB-310B region.
J.Biol.Chem., 273, 1998
|
|
6LLX
 
 | |
8H8C
 
 | | Type VI secretion system effector RhsP in its post-autoproteolysis and dimeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
