3D36
 
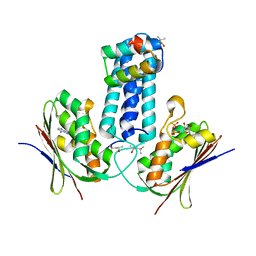 | | How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the Inhibitor Sda | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bick, M.J, Lamour, V, Rajashankar, K.R, Gordiyenko, Y, Robinson, C.V, Darst, S.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda
J.Mol.Biol., 386, 2009
|
|
7G5D
 
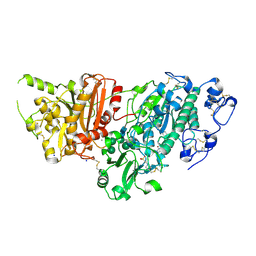 | | Crystal Structure of rat Autotaxin in complex with [(3aR,6aR)-5-(1H-benzotriazole-5-carbonyl)-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrol-2-yl]-[4-(4-chlorophenyl)phenyl]methanone, i.e. SMILES N1(C[C@@H]2[C@@H](C1)CN(C2)C(=O)c1ccc(cc1)c1ccc(cc1)Cl)C(=O)c1ccc2c(c1)N=NN2 with IC50=0.00428865 microM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3D3L
 
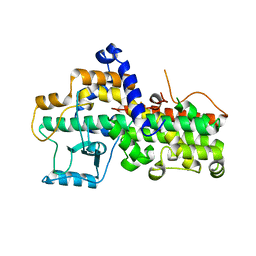 | | The 2.6 A crystal structure of the lipoxygenase domain of human arachidonate 12-lipoxygenase, 12S-type | | Descriptor: | Arachidonate 12-lipoxygenase, 12S-type, FE (III) ION | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Van Den Berg, S, Welin, M, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-12 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the lipoxygenase domain of human Arachidonate 12-lipoxygenase, 12S-type.
To be Published
|
|
3ZDF
 
 | |
7G5P
 
 | | Crystal Structure of rat Autotaxin in complex with N-[(1R,3S)-3-[(2S)-2-benzyl-4-[2-(methylamino)-2-oxoethyl]-3-oxopiperazine-1-carbonyl]cyclopentyl]-4-chloro-3-(trifluoromethyl)benzamide, i.e. SMILES C1[C@@H](CC[C@@H]1C(=O)N1CCN(C(=O)[C@@H]1Cc1ccccc1)CC(=O)NC)NC(=O)c1ccc(c(c1)C(F)(F)F)Cl with IC50=0.17269 microM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Kuhne, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G2K
 
 | | Crystal Structure of rat Autotaxin in complex with 2-chloro-5-(5-cyclopropyloxy-2-fluorophenyl)-4-[(4-methyl-5-oxo-1H-1,2,4-triazol-3-yl)methoxy]benzonitrile, i.e. SMILES c1c(ccc(c1c1cc(c(Cl)cc1OCC1=NNC(=O)N1C)C#N)F)OC1CC1 with IC50=0.0091413 microM | | Descriptor: | (1P)-4-chloro-5'-(cyclopropyloxy)-2'-fluoro-6-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methoxy][1,1'-biphenyl]-3-carbonitrile, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3CPS
 
 | | Crystal structure of Cryptosporidium parvum glyceraldehyde-3-phosphate dehydrogenase | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Cossar, D, Schapiro, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Pizarro, J, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-01 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Cryptosporidium parvum glyceraldehyde-3-phosphate dehydrogenase.
To be Published
|
|
7G31
 
 | | Crystal Structure of rat Autotaxin in complex with (3S)-3-(3-bromophenyl)-3-[[2-[2-[(3,3-difluoroazetidin-1-yl)methyl]-6-methoxyphenoxy]acetyl]amino]-N-methylpropanamide, i.e. SMILES FC1(F)CN(Cc2c(c(ccc2)OC)OCC(=O)N[C@H](c2cc(ccc2)Br)CC(=O)NC)C1 with IC50=0.276983 microM | | Descriptor: | (3R)-3-(3-bromophenyl)-3-(2-{2-[(3,3-difluoroazetidin-1-yl)methyl]-6-methoxyphenoxy}acetamido)-N-methylpropanamide, ACETATE ION, BROMIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Kuhne, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G5J
 
 | | Crystal Structure of rat Autotaxin in complex with (7E)-7-[(4-bromophenyl)methylidene]-12H-isoquinolino[2,3-a]quinazolin-5-one, i.e. SMILES N12C(=NC(=O)c3c1cccc3)/C(=C/c1ccc(cc1)Br)/c1c(C2)cccc1 with IC50=0.173306 microM | | Descriptor: | (7E,13S)-7-[(4-bromophenyl)methylidene]-7,12-dihydro-5H-isoquinolino[2,3-a]quinazolin-5-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Kumagai, Y, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
4MOK
 
 | | Pyranose 2-oxidase H167A mutant soaked with 3-fluorinated galactose (not bound) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECAETHYLENE GLYCOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
7G3F
 
 | | Crystal Structure of rat Autotaxin in complex with 2-[(2-tert-butyl-5-chloro-4-cyanophenoxy)methyl]-3-methylimidazole-4-carbonitrile, i.e. SMILES c1c(c(cc(c1C(C)(C)C)OCC1=NC=C(N1C)C#N)Cl)C#N with IC50=0.0496453 microM | | Descriptor: | 2-[(2-tert-butyl-5-chloro-4-cyanophenoxy)methyl]-1-methyl-1H-imidazole-5-carbonitrile, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Green, L, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G3U
 
 | | Crystal Structure of rat Autotaxin in complex with 5-tert-butyl-2-chloro-4-[[1-methyl-5-[5-(oxetan-3-yl)-1,2-oxazol-3-yl]imidazol-2-yl]methoxy]benzonitrile, i.e. SMILES c1c(c(cc(c1C(C)(C)C)OCC1=NC=C(N1C)C1=NOC(=C1)C1COC1)Cl)C#N with IC50=0.0167757 microM | | Descriptor: | 5-tert-butyl-2-chloro-4-({(5M)-1-methyl-5-[5-(oxetan-3-yl)-1,2-oxazol-3-yl]-1H-imidazol-2-yl}methoxy)benzonitrile, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mazunin, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3D4P
 
 | |
7G2H
 
 | | Crystal Structure of rat Autotaxin in complex with 3-fluoro-4-[1-[3-[2-[(5-methyltetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl]propanoyl]piperidin-4-yl]sulfonylbenzenesulfonamide, i.e. SMILES O=C(N1CC[C@@H](S(=O)(=O)c2c(F)cc(S(=O)(=O)N)cc2)CC1)CCc1ccc(cc1CN1N=NC(=N1)C)C(F)(F)F with IC50=0.00379624 microM | | Descriptor: | 3-fluoro-4-[1-(3-{2-[(5-methyl-2H-tetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl}propanoyl)piperidine-4-sulfonyl]benzene-1-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G43
 
 | | Crystal Structure of rat Autotaxin in complex with 3-[(6-cyclopropyl-2-methyl-1,3-benzothiazol-5-yl)oxymethyl]-4-methyl-1H-1,2,4-triazol-5-one, i.e. SMILES C(C1=NNC(=O)N1C)Oc1cc2c(cc1C1CC1)SC(=N2)C with IC50=1.05467 microM | | Descriptor: | 5-{[(6-cyclopropyl-2-methyl-1,3-benzothiazol-5-yl)oxy]methyl}-4-methyl-2,4-dihydro-3H-1,2,4-triazol-3-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
2GUG
 
 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 in complex with formate | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Formate dehydrogenase, ... | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Boiko, K.M, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2006-04-30 | | Release date: | 2006-05-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the complex of NAD-dependent formate dehydrogenase from
metylotrophic bacterium Pseudomonas sp.101 with formate.
KRISTALLOGRAFIYA, 51, 2006
|
|
7G4G
 
 | | Crystal Structure of rat Autotaxin in complex with 7-[3-(4-acetyl-3-hydroxy-2-propylphenoxy)-2-hydroxypropoxy]-4-oxo-8-propylchromene-2-carboxylic acid, i.e. SMILES c12c(c(ccc1C(=O)C=C(O2)C(=O)O)OC[C@@H](COc1c(c(c(cc1)C(=O)C)O)CCC)O)CCC with IC50=0.249971 microM | | Descriptor: | 7-[(2R)-3-(4-acetyl-3-hydroxy-2-propylphenoxy)-2-hydroxypropoxy]-4-oxo-8-propyl-4H-1-benzopyran-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Wasyliw, N, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G2P
 
 | | Crystal Structure of rat Autotaxin in complex with 1-[2-[2-cyclopropyl-6-(oxan-4-ylmethoxy)pyridine-4-carbonyl]-1,3,4,6-tetrahydropyrrolo[3,4-c]pyrrole-5-carbonyl]-4-fluoropiperidine-4-sulfonamide, i.e. SMILES F[C@@]1(S(=O)(=O)N)CCN(CC1)C(=O)N1CC2=C(CN(C(=O)c3cc(C4CC4)nc(OCC4CCOCC4)c3)C2)C1 with IC50=0.00217681 microM | | Descriptor: | 1-[5-{2-cyclopropyl-6-[(oxan-4-yl)methoxy]pyridine-4-carbonyl}-3,4,5,6-tetrahydropyrrolo[3,4-c]pyrrole-2(1H)-carbonyl]-4-fluoropiperidine-4-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
2GZX
 
 | | Crystal Structure of the TatD deoxyribonuclease MW0446 from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR237. | | Descriptor: | NICKEL (II) ION, putative TatD related DNase | | Authors: | Vorobiev, S.M, Neely, H, Seetharaman, J, Wang, D, Fang, Y, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-12 | | Release date: | 2006-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the TatD deoxyribonuclease MW0446 from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR237.
To be Published
|
|
3D4Y
 
 | | GOLGI MANNOSIDASE II complex with mannoimidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
4JA8
 
 | | Complex of Mitochondrial Isocitrate Dehydrogenase R140Q Mutant with AGI-6780 Inhibitor | | Descriptor: | 1-[5-(cyclopropylsulfamoyl)-2-thiophen-3-yl-phenyl]-3-[3-(trifluoromethyl)phenyl]urea, CALCIUM ION, GLYCEROL, ... | | Authors: | Wei, W, Chen, L, Wu, M, Jiang, F, Travins, J, Qian, K, DeLaBarre, B. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation.
Science, 340, 2013
|
|
7G4E
 
 | | Crystal Structure of rat Autotaxin in complex with (5E)-5-[(4-tert-butylphenyl)methylidene]-2-pyrrolidin-1-yl-1,3-thiazol-4-one, i.e. SMILES N1=C(S/C(=C/c2ccc(C(C)(C)C)cc2)/C1=O)N1CCCC1 with IC50=0.210793 microM | | Descriptor: | (5Z)-5-[(4-tert-butylphenyl)methylidene]-2-(pyrrolidin-1-yl)-1,3-thiazol-4(5H)-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3D5H
 
 | | Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Singh, S.B, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-05-16 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance
To be Published
|
|
7G4T
 
 | | Crystal Structure of rat Autotaxin in complex with (6-bromonaphthalen-2-yl)-[rac-(1R,2R,6S,7S)-9-(1H-benzotriazole-5-carbonyl)-4,9-diazatricyclo[5.3.0.02,6]decan-4-yl]methanone, i.e. SMILES N1(C[C@H]2[C@@H](C1)[C@H]1[C@@H]2CN(C1)C(=O)c1ccc2c(c1)N=NN2)C(=O)c1ccc2c(c1)ccc(c2)Br with IC50=0.127386 microM | | Descriptor: | CALCIUM ION, CHLORIDE ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3CRE
 
 | |
