7FRX
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 5 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-{[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]methyl}-3,4-dihydroxybenzene-1-sulfonamide, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS0
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 8 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3,4-dihydroxy-N-{2-[4-(3-hydroxybenzene-1-sulfonyl)phenyl]ethyl}benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS4
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 16 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4,4',5-trihydroxy-N-{[4-(3-hydroxybenzene-1-sulfonyl)phenyl]methyl}[1,1'-biphenyl]-2-sulfonamide, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
5RC1
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library E07a | | Descriptor: | 1-(1-methyl-1,2,3,4-tetrahydroquinolin-6-yl)methanamine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7FS8
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 21 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-[4-(3-aminobenzene-1-sulfonyl)piperazine-1-sulfonyl]benzene-1,2-diol, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
3A31
 
 | | Crystal structure of putative threonyl-tRNA synthetase ThrRS-1 from Aeropyrum pernix (selenomethionine derivative) | | Descriptor: | Probable threonyl-tRNA synthetase 1, SULFATE ION, ZINC ION | | Authors: | Shimizu, S, Juan, E.C.M, Miyashita, Y, Sato, Y, Hoque, M.M, Suzuki, K, Yogiashi, M, Tsunoda, M, Dock-Bregeon, A.-C, Moras, D, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2009-06-07 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain
J.Mol.Biol., 394, 2009
|
|
1QXL
 
 | | Crystal structure of Adenosine deaminase complexed with FR235380 | | Descriptor: | 1-((1R)-1-(HYDROXYMETHYL)-3-{6-[(5-PHENYLPENTANOYL)AMINO]-1H-INDOL-1-YL}PROPYL)-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2003-09-08 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design, synthesis, and structure-activity relationship studies of novel non-nucleoside adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
7KDB
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 35 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(4-hydroxyphenyl)-2,3-diphenyl-5-[(1H-pyrazol-3-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
3ENS
 
 | |
1AAC
 
 | | AMICYANIN OXIDIZED, 1.31 ANGSTROMS | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Cunane, L.M, Chen, Z.-W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1995-09-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | X-ray structure of the cupredoxin amicyanin, from Paracoccus denitrificans, refined at 1.31 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7KCC
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AG-270 | | Descriptor: | 1,2-ETHANEDIOL, 3-(cyclohex-1-en-1-yl)-6-(4-methoxyphenyl)-2-phenyl-5-[(pyridin-2-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, CHLORIDE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
1Y0J
 
 | | Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction | | Descriptor: | Erythroid transcription factor, ZINC ION, Zinc-finger protein ush | | Authors: | Liew, C.K, Simpson, R.J.Y, Kwan, A.H.Y, Crofts, L.A, Loughlin, F.E, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B4M
 
 | | Crystal structure of the binding protein OpuAC in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding protein | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
2ZQ7
 
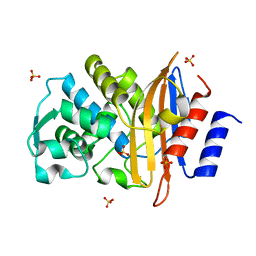 | | Apo structure of Class A beta-lactamase Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | Beta-lactamase Toho-1, SULFATE ION | | Authors: | Shimamura, T, Nitanai, Y, Uchiyama, T, Ago, H, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Improvement of the Crystal Quality by the Surface Mutations on a Beta-Lactamase Toho-1
To be Published
|
|
3I8W
 
 | |
4AD0
 
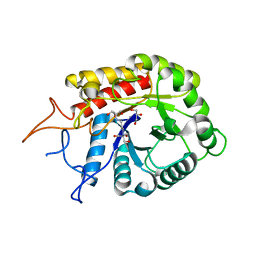 | | Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2JXA
 
 | |
3TG2
 
 | | Crystal structure of the ISC domain of VibB in complex with isochorismate | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, TRIETHYLENE GLYCOL, Vibriobactin-specific isochorismatase | | Authors: | Liu, S, Zhang, C, Niu, B, Li, N, Liu, X, Liu, M, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4J0P
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((S)-2-amino-4-methyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-01-31 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | beta-Secretase (BACE1) inhibitors with high in vivo efficacy suitable for clinical evaluation in Alzheimer's disease.
J.Med.Chem., 56, 2013
|
|
3P3H
 
 | |
3P55
 
 | |
2AWA
 
 | |
4DRN
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1S,2R)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4J1H
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((4S,6R)-2-amino-4-methyl-6-trifluoromethyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4S,6R)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-02-01 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Secretase (BACE1) Inhibitors with High In Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer s Disease
J.Med.Chem., 56, 2013
|
|
2JWE
 
 | | Solution structure of the second PDZ domain from human zonula occludens-1: A dimeric form with 3D domain swapping | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Ji, P, Wu, J.W, Zhang, J.H, Yang, Y.S, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2007-10-10 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of Zonula Occludens 1
Proteins, 79, 2011
|
|
