5O5L
 
 | |
5OIU
 
 | |
6DWO
 
 | |
4TQT
 
 | |
5CR4
 
 | |
4YY3
 
 | | 30S ribosomal subunit- HigB complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schureck, M.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2015-03-23 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | mRNA bound to the 30S subunit is a HigB toxin substrate.
Rna, 22, 2016
|
|
4TO9
 
 | | 2.0A resolution structure of BfrB (N148L) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4Z74
 
 | |
6TFL
 
 | | Lsm protein (SmAP) from Halobacterium salinarum | | Descriptor: | CALCIUM ION, GLYCEROL, RNA-binding protein Lsm, ... | | Authors: | Nikulin, A.D, Fando, M.S, Lekontseva, N.V. | | Deposit date: | 2019-11-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure and RNA-Binding Properties of Lsm Protein from Halobacterium salinarum.
Biochemistry Mosc., 86, 2021
|
|
5O9A
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L504Y-N775S) in complex with glutamate and BPAM121 at 1.78 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Rovinskaja, K, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
6THG
 
 | | Cedar Virus attachment glycoprotein (G) in complex with human ephrin-B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Pryce, R, Rissanen, I, Harlos, K, Bowden, T. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.074 Å) | | Cite: | A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus.
Life Sci Alliance, 3, 2020
|
|
6BK8
 
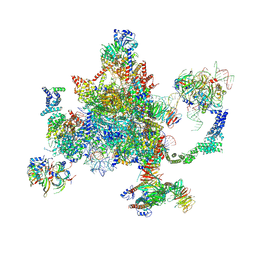 | | S. cerevisiae spliceosomal post-catalytic P complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Lea1, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2017-11-07 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the yeast spliceosomal postcatalytic P complex.
Science, 358, 2017
|
|
4Z8C
 
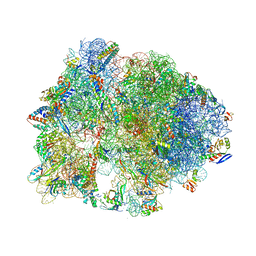 | | Crystal structure of the Thermus thermophilus 70S ribosome bound to translation inhibitor oncocin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Roy, R.N, Lomakin, I.B, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin.
Nat.Struct.Mol.Biol., 22, 2015
|
|
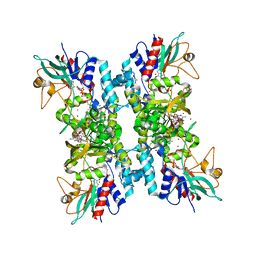
4TM1
 
 | |
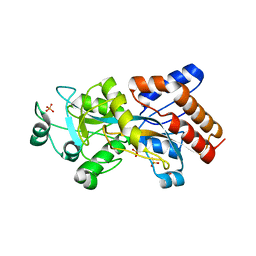
5I84
 
 | |
5IBB
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and cognate tRNAVal in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
5HAU
 
 | | Crystal structure of antimicrobial peptide Bac7(1-19) bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2015-12-30 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
5CZ4
 
 | | Yeast 20S proteasome at 2.3 A resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5D0V
 
 | |
4Z7Q
 
 | | Integrin alphaIIbbeta3 in complex with AGDV-NH2 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lin, F.-Y, Zhu, J, Springer, T.A. | | Deposit date: | 2015-04-07 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | beta-Subunit Binding Is Sufficient for Ligands to Open the Integrin alpha IIb beta 3 Headpiece.
J.Biol.Chem., 291, 2016
|
|
6TMX
 
 | | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bracher, A, Wang, H, Paul, S.S, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
5CRG
 
 | |
6JLK
 
 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6BZ6
 
 | | Thermus thermophilus 70S complex containing 16S G347U ram mutation and empty A site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2017-12-22 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding.
Nucleic Acids Res., 47, 2019
|
|
5H5U
 
 | | Mechanistic insights into the alternative translation termination by ArfA and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ma, C, Kurita, D, Li, N, Chen, Y, Himeno, H, Gao, N. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Mechanistic insights into the alternative translation termination by ArfA and RF2
Nature, 541, 2017
|
|
