168D
 
 | |
5J1O
 
 | | Excited state (Bound-like) sampled during RDC restrained Replica-averaged Metadynamics (RAM) simulations of the HIV-1 TAR complexed with cyclic peptide mimetic of Tat | | Descriptor: | Apical region (29mer) of the HIV-1 TAR element, Cyclic peptide mimetic of Tat | | Authors: | Borkar, A.N, Bardaro, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1SDS
 
 | |
9DP9
 
 | | Structure of a Tick-Borne Flavivirus xrRNA | | Descriptor: | MAGNESIUM ION, RNA (440-MER) | | Authors: | Langeberg, C.J, Kieft, J.S, Sherlock, M.E, Szucs, M.J, Vicens, Q. | | Deposit date: | 2024-09-20 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Tick-borne flavivirus exoribonuclease-resistant RNAs contain a double loop structure.
Nat Commun, 16, 2025
|
|
4ROD
 
 | | Human TFIIB-related factor 2 (Brf2) and TBP bound to TRNAU1 promoter | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Vannini, A, Gouge, J, Satia, K, Guthertz, N. | | Deposit date: | 2014-10-28 | | Release date: | 2015-12-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2.
Cell(Cambridge,Mass.), 163, 2015
|
|
2Q1R
 
 | | Crystal Structure Analysis of the RNA Dodecamer CGCGAAUUAGCG, with a G-A mismatch. | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*AP*GP*CP*G)-3') | | Authors: | Li, F, Pallan, P.S. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme.
Nucleic Acids Res., 35, 2007
|
|
1MNX
 
 | |
3PEY
 
 | | S. cerevisiae Dbp5 bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
6R9P
 
 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAUUAA RNA | | Descriptor: | AAUUAA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9O
 
 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAGGA RNA | | Descriptor: | AAGGA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.319 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1E7K
 
 | | Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment | | Descriptor: | 15.5 KD RNA BINDING PROTEIN, RNA (5'-R(*GP*CP*CP*AP*AP*UP*GP*AP*GP*GP*UP*UP*UP* AP*UP*CP*CP*GP*AP*GP*G*C(-3') | | Authors: | Vidovic, I, Nottrott, S, Harthmuth, K, Luhrmann, R, Ficner, R. | | Deposit date: | 2000-08-29 | | Release date: | 2001-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Spliceosomal 15.5Kd Protein Bound to a U4 Snrna Fragment
Mol.Cell, 6, 2000
|
|
6TZ2
 
 | | In situ structure of BmCPV RNA-dependent RNA polymerase at elongation state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-template RNA (36-MER), ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2LAC
 
 | | NMR structure of unmodified_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
3LU0
 
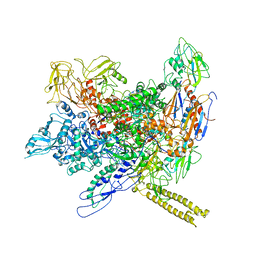 | | Molecular model of Escherichia coli core RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A. | | Deposit date: | 2010-02-16 | | Release date: | 2010-09-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.2 Å) | | Cite: | Complete structural model of Escherichia coli RNA polymerase from a hybrid approach.
Plos Biol., 8, 2010
|
|
4PDB
 
 | |
6R9M
 
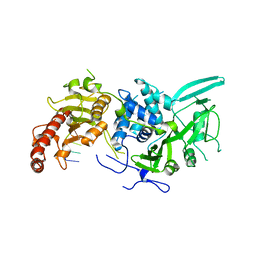 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAGGAA RNA | | Descriptor: | AAGGAA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.329 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
430D
 
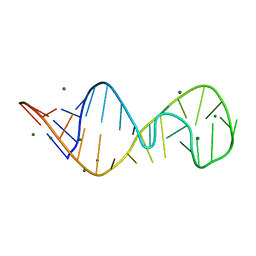 | | STRUCTURE OF SARCIN/RICIN LOOP FROM RAT 28S RRNA | | Descriptor: | MAGNESIUM ION, SARCIN/RICIN LOOP FROM RAT 28S R-RNA | | Authors: | Correll, C.C, Munishkin, A, Chan, Y.L, Ren, Z, Wool, I.G, Steitz, T.A. | | Deposit date: | 1998-10-04 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ribosomal RNA domain essential for binding elongation factors.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6R9J
 
 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with A7 RNA | | Descriptor: | A7 RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.326 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2WJ8
 
 | | Respiratory Syncitial Virus RiboNucleoProtein | | Descriptor: | BORATE ION, NUCLEOPROTEIN, RNA (5'-R(*CP*CP*CP*CP*CP*C)-3') | | Authors: | Tawar, R.G, Duquerroy, S, Vonrhein, C, Varela, P.F, Damier-Piolle, L, Castagne, N, MacLellan, K, Bedouelle, H, Bricogne, G, Bhella, D, Eleouet, J, Rey, F.A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal Structure of a Nucleocapsid-Like Nucleoprotein-RNA Complex of Respiratory Syncytial Virus
Science, 326, 2009
|
|
5XLP
 
 | |
5TEE
 
 | | Crystal structure of Gemin5 WD40 repeats in apo form | | Descriptor: | GLYCEROL, Gem-associated protein 5, SODIUM ION, ... | | Authors: | Chao, X, Tempel, W, Bian, C, Cerovina, T, He, H, Walker, J.R, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-21 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly.
Genes Dev., 30, 2016
|
|
2LA9
 
 | | NMR structure of Pseudouridine_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*(PSU)P*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
2LBQ
 
 | | NMR structure of i6A37_tyrASL | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*(6IA)P*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-04-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
5GM8
 
 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | SINEFUNGIN, tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
6TY9
 
 | | In situ structure of BmCPV RNA dependent RNA polymerase at initiation state | | Descriptor: | MAGNESIUM ION, Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*AP*AP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), RNA-dependent RNA Polymerase, ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
