2YV3
 
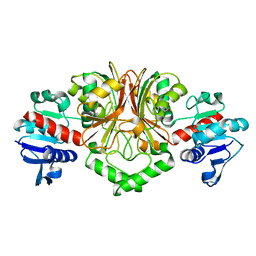 | | Crystal Structure of Aspartate Semialdehyde Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Kagawa, W, Fujikawa, N, Kurumizaka, H, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-07 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Aspartate Semialdehyde Dehydrogenase from Thermus thermophilus HB8
To be Published
|
|
4GKV
 
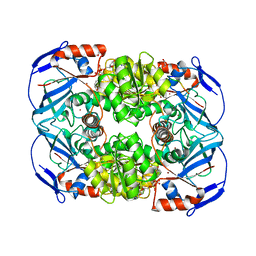 | | Structure of Escherichia coli AdhP (ethanol-inducible dehydrogenase) with bound NAD | | Descriptor: | Alcohol dehydrogenase, propanol-preferring, GLYCEROL, ... | | Authors: | Sims, P.A, Thomas, L.M, Harper, A.R, Miner, W.A, Ajufo, H.O, Branscrum, K.M, Kao, L. | | Deposit date: | 2012-08-13 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structure of Escherichia coli AdhP (ethanol-inducible dehydrogenase) with bound NAD.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2YN4
 
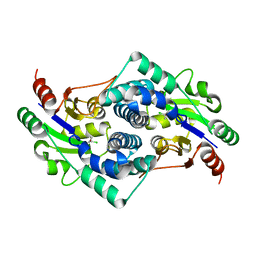 | | L-2-chlorobutryic acid bound complex L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | (2S)-2-chlorobutanoic acid, L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
4DUV
 
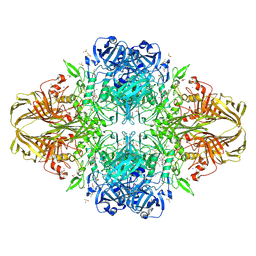 | | E. coli (lacZ) beta-galactosidase (G974A) 2-deoxy-galactosyl-enzyme and bis-Tris complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-alpha-D-galactopyranose, Beta-galactosidase, ... | | Authors: | Wheatley, R.W, Lo, S, Janzcewicz, L.J, Dugdale, M.L, Huber, R.E. | | Deposit date: | 2012-02-22 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Glucose Acceptor site of lacZ beta-galactosidase for the synthesis of allolactose - the natural inducer of the lac operon
To be Published
|
|
3VSU
 
 | | The complex structure of XylC with xylobiose | | Descriptor: | Xylosidase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
2BXV
 
 | | Dual binding mode of a novel series of DHODH inhibitors | | Descriptor: | 2-({[3-FLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)CYCLOPENT-1-ENE-1-CARBOXYLIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Baumgartner, R, Walloschek, M, Karlik, M, Gotschlich, A, Tasler, S, Mies, J, Leban, J. | | Deposit date: | 2005-07-27 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J. Med. Chem., 49, 2006
|
|
1OM0
 
 | | crystal structure of xylanase inhibitor protein (XIP-I) from wheat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Xylanase Inhibitor Protein I | | Authors: | Payan, F, Flatman, R, Porciero, S, Williamson, G, Juge, N, Roussel, A. | | Deposit date: | 2003-02-24 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson).
Biochem.J., 372, 2003
|
|
4H7M
 
 | |
2YDG
 
 | | Ascorbate co-crystallized HEWL. | | Descriptor: | ASCORBIC ACID, Lysozyme C, SODIUM ION | | Authors: | De la Mora, E, Carmichael, I, Garman, E.F. | | Deposit date: | 2011-03-19 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effective Scavenging at Cryotemperatures: Further Increasing the Dose Tolerance of Protein Crystals.
J.Synchrotron.Radiat., 18, 2011
|
|
4H9C
 
 | | Radiation damage study of lysozyme - 0.77 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1998 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H9I
 
 | | Radiation damage study of lysozyme - 1.05 MGy | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Sutton, K.A, Snell, E.H. | | Deposit date: | 2012-09-24 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2002 Å) | | Cite: | Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H2G
 
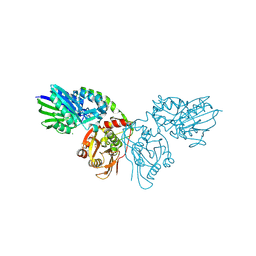 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with adenosine | | Descriptor: | 5'-nucleotidase, ADENOSINE, CALCIUM ION, ... | | Authors: | Straeter, N, Knapp, K.M, Zebisch, M, Pippel, J. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
1OO6
 
 | | Nitroreductase from e-coli in complex with the dinitrobenzamide prodrug SN23862 | | Descriptor: | 5-[BIS-2(CHLORO-ETHYL)-AMINO]-2,4-DINTRO-BENZAMIDE, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-03 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
2BER
 
 | | Y370G Active Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). | | Descriptor: | BACTERIAL SIALIDASE, N-acetyl-beta-neuraminic acid, SODIUM ION | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G.L. | | Deposit date: | 2004-11-30 | | Release date: | 2005-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Mechanism of Action of an Inverting Mutant Sialidase.
Biochemistry, 44, 2005
|
|
3W1L
 
 | | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-chloroorotate | | Descriptor: | 5-chloro-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-chloroorotate
TO BE PUBLISHED
|
|
2Y99
 
 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356 complex with co-enzyme NAD | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-12 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3VWO
 
 | |
3W1X
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-121 | | Descriptor: | 5-{2-[4-(cyanomethyl)phenyl]ethyl}-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-22 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-121.
To be Published
|
|
1OFE
 
 | |
5P9Z
 
 | | humanized rat COMT in complex with 6-(4-fluorophenyl)quinazolin-8-ol | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 6-(4-fluorophenyl)quinazolin-8-ol, CHLORIDE ION, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-10-19 | | Release date: | 2017-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a COMT complex
To be published
|
|
4H5U
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 | | Descriptor: | CACODYLATE ION, GLYCEROL, Probable hydrolase NIT2 | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2Z19
 
 | |
4HGO
 
 | |
1X39
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-3-(ANILINOMETHYL)-5,6,7,8-TETRAHYDRO-5-(HYDROXYMETHYL)-IMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Hrmova, M, Streltsov, V.A, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2005-05-02 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationale for low-nanomolar binding of transition state mimics to a family GH3 beta-D-glucan glucohydrolase from barley.
Biochemistry, 44, 2005
|
|
2ZB4
 
 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADP and 15-keto-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2, ... | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
