9BXQ
 
 | |
9BXO
 
 | |
9BXI
 
 | |
9BW0
 
 | | RNA Polymerase II - No ATP | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Calero, G. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BVT
 
 | | RNA Pol II - High Mn(+2) concentration | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Calero, G. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BU1
 
 | |
9BTS
 
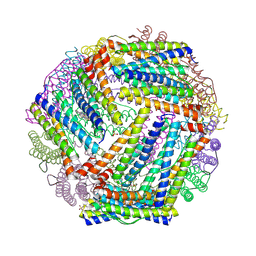 | | Crystal structure of the bacterioferritin (Bfr) and ferritin (Ftn) heterooligomer complex from Acinetobacter baumannii | | Descriptor: | Bacterioferritin (Bfr), Ferritin (Ftn), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of Acinetobacter baumannii bacterioferritin reveals a heteropolymer of bacterioferritin and ferritin subunits.
Sci Rep, 14, 2024
|
|
9BRX
 
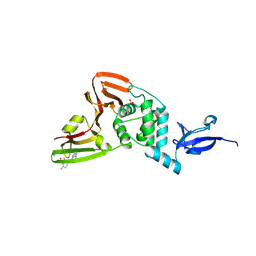 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 10 | | Descriptor: | (4R)-N-(2,4-dimethylphenyl)-7-methyl[1,2,4]triazolo[4,3-a]pyrimidin-5-amine, Papain-like protease nsp3, SULFATE ION, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9BRW
 
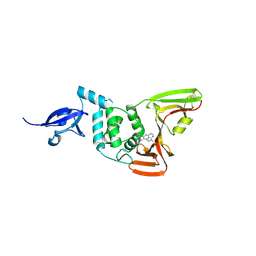 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 7 | | Descriptor: | CHLORIDE ION, N-(2-chlorophenyl)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9BRV
 
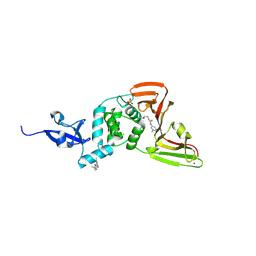 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 5 | | Descriptor: | CHLORIDE ION, N-[2-(dimethylamino)ethyl]-N'-(3-methylphenyl)thiourea, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9BQV
 
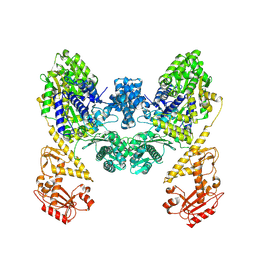 | | DdmD dimer apoprotein | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
9BQJ
 
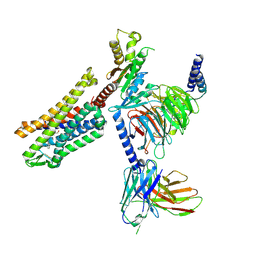 | | RO76 bound muOR-Gi1-scFv16 complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Majumdar, S, Kobilka, B.K. | | Deposit date: | 2024-05-10 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling Modulation Mediated by Ligand Water Interactions with the Sodium Site at mu OR.
Acs Cent.Sci., 10, 2024
|
|
9BQE
 
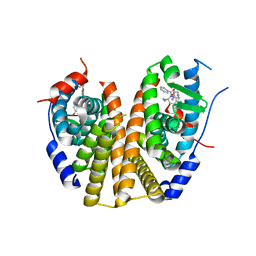 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with (6'-hydroxy-1'-(4-(2-(1-propylpyrrolidin-3-yl)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-(4-{[(3R)-1-propylpyrrolidin-3-yl]methoxy}phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Young, K.Y, Fanning, S.W. | | Deposit date: | 2024-05-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with (6'-hydroxy-1'-(4-(2-(1-propylpyrrolidin-3-yl)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone
To Be Published
|
|
9BPF
 
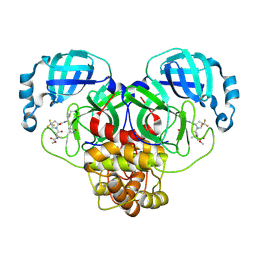 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
9BOO
 
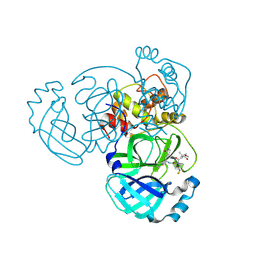 | | Crystal structure of MERS-CoV Nsp5 in complex with PF-07817883 | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
9BNR
 
 | | 4-(2-isothiocyanatoethyl)benzenesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | Macrophage migration inhibitory factor, N-[2-(4-sulfamoylphenyl)ethyl]methanethioamide, SULFATE ION | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
9BNQ
 
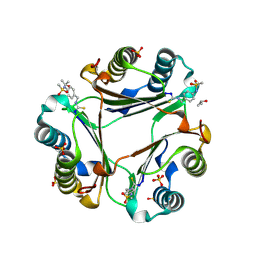 | | N-(4-(isothiocyanatomethyl)phenyl)methanesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, N-{[4-(methanesulfonamido)phenyl]methyl}methanethioamide, ... | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
9BNA
 
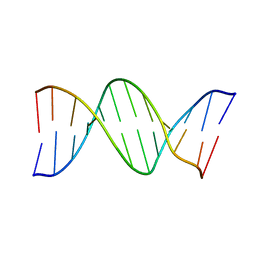 | |
9BN9
 
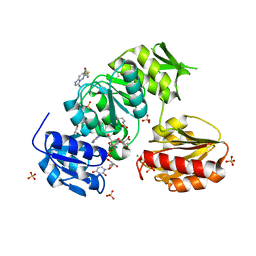 | |
9BN8
 
 | |
9BLN
 
 | | The structure of human Pdcd4 bound to the 40S-eIF4A-eIF3-eIF1 complex | | Descriptor: | 40S ribosomal protein S14, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Yifei, D, Albacete-Albacete, L, Ramakrishnan, V, S Fraser, C. | | Deposit date: | 2024-04-30 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
9BLG
 
 | | Crystal structure of non-receptor protein tyrosine phosphatase SHP2 in complex with PF-07284892 | | Descriptor: | (1S)-1'-{6-[(2-amino-3-chloropyridin-4-yl)sulfanyl]-1,2,4-triazin-3-yl}-1,3-dihydrospiro[indene-2,4'-piperidin]-1-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Bester, S.M, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | SHP2 Inhibition Sensitizes Diverse Oncogene-Addicted Solid Tumors to Re-treatment with Targeted Therapy.
Cancer Discov, 13, 2023
|
|
9BKY
 
 | |
9BKO
 
 | | DHODH in complex with Ligand 26 | | Descriptor: | (2P,6P)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-2-(2-methylphenyl)-4-[(2R)-1,1,1-trifluoropropan-2-yl]isoquinolin-1(2H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of JNJ-74856665: A Novel Isoquinolinone DHODH Inhibitor for the Treatment of AML.
J.Med.Chem., 67, 2024
|
|
9BKN
 
 | | DHODH in complex with Ligand 16 | | Descriptor: | (2P,6P)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-2-(2-methylphenyl)-4-(propan-2-yl)isoquinolin-1(2H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of JNJ-74856665: A Novel Isoquinolinone DHODH Inhibitor for the Treatment of AML.
J.Med.Chem., 67, 2024
|
|
