3WN6
 
 | | Crystal structure of alpha-amylase AmyI-1 from Oryza sativa | | Descriptor: | Alpha-amylase, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Ochiai, A, Sugai, H, Harada, K, Tanaka, S, Ishiyama, Y, Ito, K, Tanaka, T, Uchiumi, T, Taniguchi, M, Mitsui, T. | | Deposit date: | 2013-12-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of alpha-amylase from Oryza sativa: molecular insights into enzyme activity and thermostability
Biosci.Biotechnol.Biochem., 78, 2014
|
|
1ZYK
 
 | | Anthranilate Phosphoribosyltransferase in complex with PRPP, anthranilate and magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-AMINOBENZOIC ACID, Anthranilate phosphoribosyltransferase, ... | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
3OLC
 
 | |
4FEW
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor pyrazolopyrimidine PP2 | | Descriptor: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4JCE
 
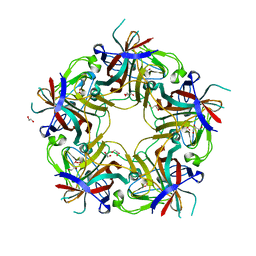 | | L54F Variant of JC Polyomavirus Major Capsid Protein VP1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1 | | Authors: | Stehle, T, Stroh, L.J. | | Deposit date: | 2013-02-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Progressive multifocal leukoencephalopathy-associated mutations in the JC polyomavirus capsid disrupt lactoseries tetrasaccharide c binding.
MBio, 4, 2013
|
|
3GN1
 
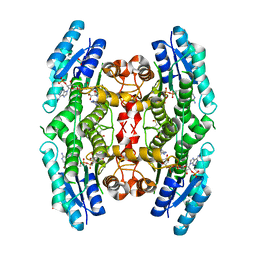 | | Structure of Pteridine Reductase 1 (PTR1) from TRYPANOSOMA BRUCEI in ternary complex with cofactor (NADP+) and inhibitor (DDD00067116) | | Descriptor: | 1H-benzimidazol-2-amine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Brenk, R, Hunter, W.N. | | Deposit date: | 2009-03-16 | | Release date: | 2009-12-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment hits discovered by virtual screening.
J.Med.Chem., 52, 2009
|
|
4C0M
 
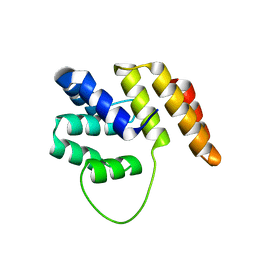 | | Crystal Structure of the N terminal domain of wild type TRIF (TIR- domain-containing adapter-inducing interferon-beta) | | Descriptor: | TIR DOMAIN-CONTAINING ADAPTER MOLECULE 1 | | Authors: | Ullah, M.O, Ve, T, Mangan, M, Alaidarous, M, Sweet, M.J, Mansell, A, Kobe, B. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Tlr Signalling Adaptor Trif/Ticam-1 Has an N-Terminal Helical Domain with Structural Similarity to Ifit Proteins
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4C1C
 
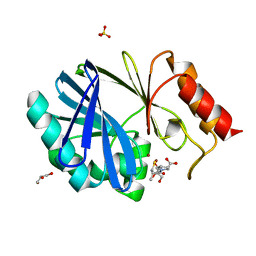 | | Crystal structure of the metallo-beta-lactamase BCII with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, BETA-LACTAMASE 2, GLYCEROL, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
5RF9
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z217038356 | | Descriptor: | 1-[(2~{S})-2-methylmorpholin-4-yl]-2-pyrazol-1-yl-ethanone, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3GYD
 
 | |
4JE5
 
 | | Crystal structure of the aromatic aminotransferase Aro8, a putative alpha-aminoadipate aminotransferase in Saccharomyces cerevisiae | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic/aminoadipate aminotransferase 1, ... | | Authors: | Bulfer, S.L, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2013-02-26 | | Release date: | 2013-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae Aro8, a putative alpha-aminoadipate aminotransferase.
Protein Sci., 22, 2013
|
|
5R4X
 
 | | XChem fragment screen -- CRYSTAL STRUCTURE OF THE BROMODOMAIN OF THE HUMAN ATAD2 in complex with N13413a | | Descriptor: | 1,2-ETHANEDIOL, 4-acetyl-N-ethylpiperazine-1-carboxamide, ATPase family AAA domain-containing protein 2, ... | | Authors: | Talon, R, Krojer, T, Fairhead, M, Sethi, R, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Wright, N, MacLean, E, Zhang, R, Dias, A, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2020-02-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | XChem fragment screen
To Be Published
|
|
3QAT
 
 | |
1QNS
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3QHX
 
 | |
7KZB
 
 | |
2GO2
 
 | |
3TZU
 
 | |
1DR9
 
 | | CRYSTAL STRUCTURE OF A SOLUBLE FORM OF B7-1 (CD80) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T LYMPHOCYTE ACTIVATION ANTIGEN | | Authors: | Ikemizu, S, Jones, E.Y, Stuart, D.I, Davis, S.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and dimerization of a soluble form of B7-1.
Immunity, 12, 2000
|
|
3U2D
 
 | | S. aureus GyrB ATPase domain in complex with small molecule inhibitor | | Descriptor: | 4-bromo-5-methyl-N-[1-(3-nitropyridin-2-yl)piperidin-4-yl]-1H-pyrrole-2-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
4CDF
 
 | | Human DPP1 in complex with (2S,4S)-N-((1S)-1-cyano-2-(4-(4- cyanophenyl)phenyl)ethyl)-4-hydroxy-piperidine-2-carboxamide | | Descriptor: | (2S,4S)-N-[(2S)-1-azanylidene-3-[4-(4-cyanophenyl)phenyl]propan-2-yl]-4-oxidanyl-piperidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Debreczeni, J, Edman, K, Furber, M, Tiden, A, Gardiner, P, Mete, T, Ford, R, Millichip, I, Stein, L, Mather, A, Kinchin, E, Luckhurst, C, Cage, P, Sanghanee, H, Breed, J, Wissler, L. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cathepsin C Inhibitors: Property Optimization and Identification of a Clinical Candidate.
J.Med.Chem., 57, 2014
|
|
4C4A
 
 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SAH | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, PROTEIN ARGININE N-METHYLTRANSFERASE 7, ... | | Authors: | Cura, V, Troffer-Charlier, N, Bonnefond, L, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-09-02 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight Into Arginine Methylation by the Mouse Protein Arginine Methyltransferase 7: A Zinc Finger Freezes the Mimic of the Dimeric State Into a Single Active Site.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7KUS
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 10 H137A Mutant in Complex with N8-Acetylspermidine (Tetrahedral Intermediate) | | Descriptor: | 1,2-ETHANEDIOL, 1-({4-[(3-aminopropyl)amino]butyl}amino)ethane-1,1-diol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-11-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Crystallographic Snapshots of Substrate Binding in the Active Site of Histone Deacetylase 10.
Biochemistry, 60, 2021
|
|
7KVI
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sevrioukova, I. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength.
Int J Mol Sci, 22, 2021
|
|
4JF5
 
 | | Structure of OXA-23 at pH 4.1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Basis for Carbapenemase Activity of the OXA-23 beta-Lactamase from Acinetobacter baumannii.
Chem.Biol., 20, 2013
|
|
