4D72
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCOSIDE HYDROLASE, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D88
 
 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BXQ490 | | Descriptor: | (3S,4S,5R)-3-{4-amino-3-fluoro-5-[(2S)-3,3,3-trifluoro-2-hydroxypropyl]benzyl}-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
1V3X
 
 | | Factor Xa in complex with the inhibitor 1-[6-methyl-4,5,6,7-tetrahydrothiazolo(5,4-c)pyridin-2-yl] carbonyl-2-carbamoyl-4-(6-chloronaphth-2-ylsulphonyl)piperazine | | Descriptor: | (2R)-4-[(6-CHLORO-2-NAPHTHYL)SULFONYL]-1-[(5-METHYL-4,5,6,7-TETRAHYDRO[1,3]THIAZOLO[5,4-C]PYRIDIN-2-YL)CARBONYL]PIPERAZ INE-2-CARBOXAMIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and conformational analysis of a non-amidine factor Xa inhibitor that incorporates 5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridine as S4 binding element
J.Med.Chem., 47, 2004
|
|
2F47
 
 | | Xray crystal structure of T4 lysozyme mutant L20/R63A liganded to methylguanidinium | | Descriptor: | 1-METHYLGUANIDINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-22 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
7G5Z
 
 | | Crystal Structure of rat Autotaxin in complex with N-(5-chloro-2,3-dihydro-1H-inden-2-yl)-6-(5-methylpyrimidin-2-yl)-5,7-dihydropyrrolo[3,4-d]pyrimidin-2-amine, i.e. SMILES Clc1cc2c(cc1)C[C@@H](C2)Nc1ncc2CN(Cc2n1)c1ncc(cn1)C with IC50=0.0693825 microM | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Pinard, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G56
 
 | | Crystal Structure of rat Autotaxin in complex with a dimer of [(2R,4S,5S)-5-ethynyl-1-azabicyclo[2.2.2]octan-2-yl]methyl N-cyclohexylcarbamate, i.e. SMILES O=C(NC1CCCCC1)OC[C@H]2C[C@@H]3CCN2C[C@@H]3C#CC#C[C@H]4CN5CC[C@H]4C[C@@H]5COC(=O)NC6CCCCC6 with IC50=4.47875 microM | | Descriptor: | (R)-(cyclohexylamino){3-[(3R,4R)-3-{4-[(1R,3R,4R,6R)-6-{[(R)-(cyclohexylamino)(hydroxy)methoxy]methyl}-1-azabicyclo[2.2.2]octan-3-yl]butyl}piperidin-4-yl]propoxy}methanol, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G6F
 
 | | Crystal Structure of rat Autotaxin in complex with (3R)-3-[[2-[6-(4-acetylpiperazin-1-yl)-4-oxoquinazolin-3-yl]acetyl]amino]-N-methyl-3-(4-nitrophenyl)propanamide, i.e. SMILES c1c(ccc(c1)[C@@H](CC(=O)NC)NC(=O)CN1C=Nc2ccc(cc2C1=O)N1CCN(CC1)C(=O)C)N(=O)=O with IC50=0.121117 microM | | Descriptor: | (3S)-3-{2-[6-(4-acetylpiperazin-1-yl)-4-oxoquinazolin-3(4H)-yl]acetamido}-N-methyl-3-(4-nitrophenyl)propanamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Martin-Rainer, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3ZTX
 
 | | Aurora kinase selective inhibitors identified using a Taxol-induced checkpoint sensitivity screen. | | Descriptor: | 2-((4-(4-HYDROXYPIPERIDIN-1-YL)PHENYL)AMINO)-5,11-DIMETHYL-5H-BENZO[E]PYRIMIDO [5,4-B][1,4]DIAZEPIN-6(11H)-ONE, INNER CENTROMERE PROTEIN A, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | Kwiatkowski, N, Villa, F, Musacchio, A, Gray, N. | | Deposit date: | 2011-07-12 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Aurora Kinase Inhibitors Identified Using a Taxol- Induced Checkpoint Sensitivity Screen.
Acs Chem.Biol., 7, 2012
|
|
6MNV
 
 | | Crystal structure of X. citri phosphoglucomutase in complex with CH2FG1P | | Descriptor: | 1-deoxy-1-fluoro-2-O-phosphono-alpha-D-gluco-hept-2-ulopyranose, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase, ... | | Authors: | Beamer, L, Stiers, K. | | Deposit date: | 2018-10-03 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitory Evaluation of alpha PMM/PGM fromPseudomonas aeruginosa: Chemical Synthesis, Enzyme Kinetics, and Protein Crystallographic Study.
J.Org.Chem., 84, 2019
|
|
2WSU
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre | | Descriptor: | GLYCEROL, NITRATE ION, PUTATIVE FIBER PROTEIN | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
7KA0
 
 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and phenylalanine | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Wower, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
1VA5
 
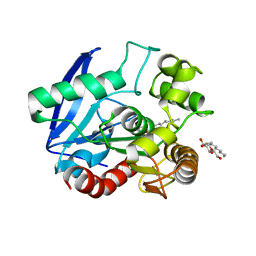 | | Antigen 85C with octylthioglucoside in active site | | Descriptor: | Antigen 85-C, octyl 1-thio-beta-D-glucopyranoside | | Authors: | Ronning, D.R, Vissa, V, Besra, G.S, Belisle, J.T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-11 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mycobacterium tuberculosis Antigen 85A and 85C Structures Confirm Binding Orientation and Conserved Substrate Specificity
J.Biol.Chem., 279, 2004
|
|
7K98
 
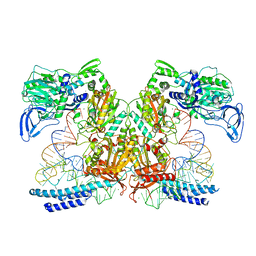 | | Preaminoacylation complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine (F-AMS) | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
1VAK
 
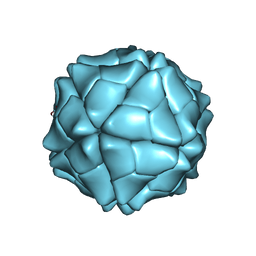 | | T=1 capsid structure of Sesbania mosaic virus coat protein deletion mutant CP-N(delta)65 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumer, P.S, Vijay, C.S, Saravanan, V, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2004-02-18 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly
J.Mol.Biol., 342, 2004
|
|
1VB4
 
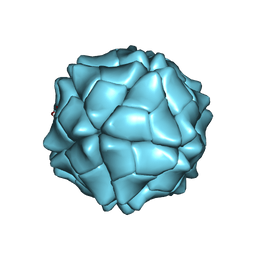 | | T=1 capsid structure of Sesbania mosaic virus coat protein deletion mutant CP-N(delta)36 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Sangita, V, Lokesh, G.L, Satheshkumar, P.S, Vijay, C.S, Saravanan, V, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2004-02-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly
J.Mol.Biol., 342, 2004
|
|
3FZ5
 
 | | Crystal structure of possible 2-hydroxychromene-2-carboxylate isomerase from Rhodobacter sphaeroides | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Chang, C, Hatzos, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of possible 2-hydroxychromene-2-carboxylate isomerase from Rhodobacter sphaeroides
To be Published
|
|
3MHJ
 
 | | Human tankyrase 2 - catalytic PARP domain in complex with 1-methyl-3-(trifluoromethyl)-5h-benzo[c][1,8]naphtyridine-6-one | | Descriptor: | 1-methyl-3-(trifluoromethyl)benzo[c][1,8]naphthyridin-6(5H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Markova, N, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
5R4V
 
 | | XChem fragment screen -- CRYSTAL STRUCTURE OF THE BROMODOMAIN OF THE HUMAN ATAD2 in complex with N13475a | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Talon, R, Krojer, T, Fairhead, M, Sethi, R, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Wright, N, MacLean, E, Zhang, R, Dias, A, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2020-02-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | XChem fragment screen
To Be Published
|
|
3QEU
 
 | | The crystal structure of TCR DMF5 | | Descriptor: | DMF5 alpha chain, DMF5 beta chain, GLYCEROL, ... | | Authors: | Borbulevych, O.Y, Santhanagopolan, S.M, Baker, B.M. | | Deposit date: | 2011-01-20 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | TCRs Used in Cancer Gene Therapy Cross-React with MART-1/Melan-A Tumor Antigens via Distinct Mechanisms.
J.Immunol., 187, 2011
|
|
1US3
 
 | | Native xylanase10C from Cellvibrio japonicus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-1,4-XYLANASE PRECURSOR, GLYCEROL, ... | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1K33
 
 | | Crystal structure analysis of the gp41 core mutant | | Descriptor: | Transmembrane glycoprotein GP41 | | Authors: | Shu, W, Lu, M. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion.
Biochemistry, 41, 2002
|
|
3MNZ
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide bearing Ala substitutions in a helical conformation | | Descriptor: | ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, SODIUM ION, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
3QK0
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | Descriptor: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
1YCP
 
 | |
3Q7L
 
 | |
