5LNK
 
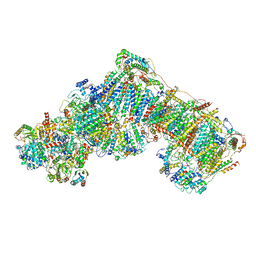 | | Entire ovine respiratory complex I | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 4'-PHOSPHOPANTETHEINE, ... | | Authors: | Fiedorczuk, K, Letts, J.A, Kaszuba, K, Sazanov, L.A. | | Deposit date: | 2016-08-04 | | Release date: | 2016-11-23 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the entire mammalian mitochondrial complex I.
Nature, 538, 2016
|
|
4YGH
 
 | |
4Y91
 
 | |
3RKX
 
 | |
3RIR
 
 | |
3RKW
 
 | |
3PCU
 
 | | Crystal structure of human retinoic X receptor alpha ligand-binding domain complexed with LX0278 and SRC1 peptide | | Descriptor: | 2-[(2S)-6-(2-methylbut-3-en-2-yl)-7-oxo-2,3-dihydro-7H-furo[3,2-g]chromen-2-yl]propan-2-yl acetate, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Zhang, Y, Shen, H, Chen, J, Li, C, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-10-22 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (+)-Rutamarin as a Dual Inducer of Both GLUT4 Translocation and Expression Efficiently Ameliorates Glucose Homeostasis in Insulin-Resistant Mice.
Plos One, 7, 2012
|
|
3PEQ
 
 | | PPARd complexed with a phenoxyacetic acid partial agonist | | Descriptor: | IODIDE ION, Peroxisome proliferator-activated receptor delta, [(4-{butyl[2-methyl-4'-(methylsulfanyl)biphenyl-3-yl]sulfamoyl}naphthalen-1-yl)oxy]acetic acid, ... | | Authors: | Lambert, M.H, Evans, K.A, Shearer, B.G, Wisnoski, D.D, Shi, D, Jin, J, Rivero, R.A, Sparks, S.M, Winegar, D.A, Billin, A.N, Britt, C, Way, J.M, Leesnitzer, L.M, Merrihew, R.V. | | Deposit date: | 2010-10-27 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phenoxyacetic acid PPARd partial agonists for the treatment of type 2 diabetes: synthesis, optimization, and in vivo efficacy
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5A5P
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 8-2-(dimethylamino)ethylamino-3-methyl-1,2-dihydroquinolin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 8-{[2-(dimethylamino)ethyl]amino}-3-methyl-1,2-dihydroquinolin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-06-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-Based Discovery of Low-Micromolar Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
5A81
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 8-(3R,4R) -3-(cyclohexylmethoxy)piperidin-4-yl-amino-3-methyl-1,2-dihydro-1,7- naphthyridin-2-one | | Descriptor: | (3R,4R)-3-(cyclohexylmethoxy)piperidin-4-yl]amino}-3-methyl-1,2-dihydro-1,7-naphthyridin-2-one, 1,2-ETHANEDIOL, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-07-11 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Optimization of Naphthyridones Into Potent Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
2H0N
 
 | |
3R8D
 
 | |
3VUX
 
 | | Crystal structure of A20 ZF7 in complex with linear ubiquitin, form II | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, Polyubiquitin-C, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
3ULF
 
 | |
3R29
 
 | | Crystal structure of RXRalpha ligand-binding domain complexed with corepressor SMRT2 | | Descriptor: | Nuclear receptor corepressor 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Chen, L, Chen, J, Jiang, H, Shen, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for retinoic x receptor repression on the tetramer.
J.Biol.Chem., 286, 2011
|
|
3UE6
 
 | |
3R2A
 
 | | Crystal structure of RXRalpha ligand-binding domain complexed with corepressor SMRT2 and antagonist rhein | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Nuclear receptor corepressor 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Chen, L, Chen, J, Jiang, H, Shen, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for retinoic x receptor repression on the tetramer.
J.Biol.Chem., 286, 2011
|
|
3R8A
 
 | |
3OLS
 
 | | Crystal structure of estrogen receptor beta ligand binding domain | | Descriptor: | ESTRADIOL, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, Rose, R, Carraz, M, Visser, A, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-26 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and crystal structure of a phosphorylated estrogen receptor ligand binding domain.
Chembiochem, 11, 2010
|
|
3VUY
 
 | | Crystal structure of A20 ZF7 in complex with linear tetraubiquitin | | Descriptor: | POTASSIUM ION, Polyubiquitin-C, Tumor necrosis factor alpha-induced protein 3, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
2GDG
 
 | |
3OMQ
 
 | | Fragment-Based Design of novel Estrogen Receptor Ligands | | Descriptor: | 2-[(trifluoromethyl)sulfonyl]-1,2,3,4-tetrahydroisoquinolin-6-ol, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, van Otterlo, W.A, Rose, R, Fuchs, S, Dominguez Seoane, M, Waldmann, H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design and Evaluation of Fragment-Like Estrogen Receptor Tetrahydroisoquinoline Ligands from a Scaffold-Detection Approach.
J.Med.Chem., 54, 2011
|
|
3FWC
 
 | | Sac3:Sus1:Cdc31 complex | | Descriptor: | Cell division control protein 31, Nuclear mRNA export protein SAC3, Protein SUS1, ... | | Authors: | Stewart, M, Jani, D. | | Deposit date: | 2009-01-17 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Mol.Cell, 33, 2009
|
|
4C7Q
 
 | |
3IXP
 
 | |
