4JJK
 
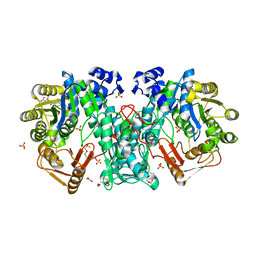 | | Crystal Structure of N10-Formyltetrahydrofolate Synthetase with Folate | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, FOLIC ACID, ... | | Authors: | Celeste, L.R, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2013-03-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of N10-formyltetrahydrofolate synthetase derived from complexes with intermediates and inhibitors.
Protein Sci., 21, 2012
|
|
4Y86
 
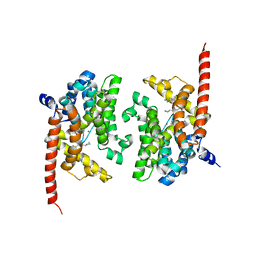 | | Crystal structure of PDE9 in complex with racemic inhibitor C33 | | Descriptor: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, 6-{[(1S)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | Authors: | Huang, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
2OKE
 
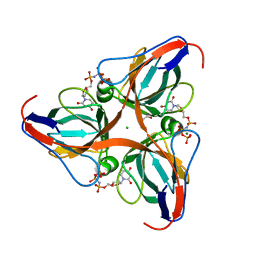 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4FAD
 
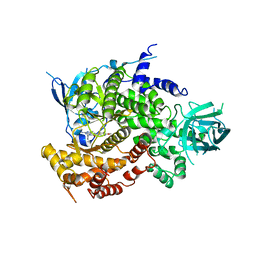 | | Design and Synthesis of a Novel Pyrrolidinyl Pyrido Pyrimidinone Derivative as a Potent Inhibitor of PI3Ka and mTOR | | Descriptor: | 2-amino-6-(5-fluoro-6-methoxypyridin-3-yl)-4-methyl-8-(pyrrolidin-1-yl)pyrido[2,3-d]pyrimidin-7(8H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Greasley, S.E, Knighton, D.R, LaFleur Rogers, C.M. | | Deposit date: | 2012-05-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and synthesis of a novel pyrrolidinyl pyrido pyrimidinone derivative as a potent inhibitor of PI3Kalpha and mTOR
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7FXI
 
 | | Crystal Structure of human FABP4 in complex with rac-(1R,3S)-2,2-dimethyl-3-[(2-phenylphenyl)carbamoyl]cyclopropane-1-carboxylic acid, i.e. SMILES C(=O)(O)[C@@H]1[C@H](C(=O)Nc2ccccc2c2ccccc2)C1(C)C with IC50=0.582 microM | | Descriptor: | (1S,3R)-3-[([1,1'-biphenyl]-2-yl)carbamoyl]-2,2-dimethylcyclopropane-1-carboxylic acid, CHLORIDE ION, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3O6H
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, 2-[({4-[(ethylamino)methyl]-3-(trifluoromethyl)-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2NW0
 
 | | Crystal structure of a lysin | | Descriptor: | ACETATE ION, PlyB | | Authors: | Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A Crystal Structure of the Catalytic Domain of PlyB, a Bacteriophage Lysin Active Against Bacillus anthracis.
J.Mol.Biol., 366, 2007
|
|
6MNV
 
 | | Crystal structure of X. citri phosphoglucomutase in complex with CH2FG1P | | Descriptor: | 1-deoxy-1-fluoro-2-O-phosphono-alpha-D-gluco-hept-2-ulopyranose, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase, ... | | Authors: | Beamer, L, Stiers, K. | | Deposit date: | 2018-10-03 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitory Evaluation of alpha PMM/PGM fromPseudomonas aeruginosa: Chemical Synthesis, Enzyme Kinetics, and Protein Crystallographic Study.
J.Org.Chem., 84, 2019
|
|
5KZC
 
 | | Crystal structure of an HIV-1 gp120 engineered outer domain with a Man9 glycan at position N276, in complex with broadly neutralizing antibody VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Engineered outer domain of gp120, VRC01 Fab heavy chain, ... | | Authors: | Julien, J.-P, Jardine, J.G, Diwanji, D, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-07-24 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
3NWX
 
 | | X-ray structure of ester chemical analogue [O-Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-12 | | Release date: | 2011-11-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TI1
 
 | | CDK2 in complex with SUNITINIB | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Alam, R, Schonbrunn, E. | | Deposit date: | 2011-08-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
4TT0
 
 | |
6G01
 
 | |
1XRB
 
 | | S-adenosylmethionine synthetase (MAT, ATP: L-methionine S-adenosyltransferase, E.C.2.5.1.6) in which MET residues are replaced with selenomethionine residues (MSE) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Takusagawa, F, Kamitori, S, Misaki, S, Markham, G.D. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of S-adenosylmethionine synthetase.
J.Biol.Chem., 271, 1996
|
|
3ECN
 
 | | Crystal structure of PDE8A catalytic domain in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
4DJY
 
 | |
5L8R
 
 | | The structure of plant photosystem I super-complex at 2.6 angstrom resolution. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Mazor, Y, Borovikova, A, Caspy, I, Nelson, N. | | Deposit date: | 2016-06-08 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the plant photosystem I supercomplex at 2.6 angstrom resolution.
Nat Plants, 3, 2017
|
|
7G05
 
 | | Crystal Structure of human FABP4 in complex with 2,6-dichloro-4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenol, i.e. SMILES C(C(F)(F)F)(C(F)(F)F)(c1cc(c(c(c1)Cl)O)Cl)O with IC50=12 microM | | Descriptor: | 2,6-dichloro-4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenol, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3A1B
 
 | | Crystal structure of the DNMT3A ADD domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, Histone H3.1, ... | | Authors: | Otani, J, Arita, K, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2009-03-28 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain
Embo Rep., 10, 2009
|
|
5A8O
 
 | | Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with cellotetraose | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
2MK3
 
 | | Solution NMR structure of gp41 ectodomain monomer on a DPC micelle | | Descriptor: | Transmembrane glycoprotein, chimeric construct | | Authors: | Roche, J, Louis, J.M, Grishaev, A, Ying, J, Bax, A. | | Deposit date: | 2014-01-23 | | Release date: | 2014-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dissociation of the trimeric gp41 ectodomain at the lipid-water interface suggests an active role in HIV-1 Env-mediated membrane fusion.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4AG4
 
 | | Crystal structure of a DDR1-Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1, ... | | Authors: | Carafoli, F, Mayer, M.C, Shiraishi, K, Pecheva, M.A, Chan, L.Y, Nan, R, Leitinger, B, Hohenester, E. | | Deposit date: | 2012-01-24 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Discoidin Domain Receptor 1 Extracellular Region Bound to an Inhibitory Fab Fragment Reveals Features Important for Signaling.
Structure, 20, 2012
|
|
1HXC
 
 | | CRYSTAL STRUCTURE OF TEAS C440W | | Descriptor: | 1-HYDROXY-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENE PHOSPHONIC ACID, 5-EPI-ARISTOLOCHENE SYNTHASE | | Authors: | Starks, C.S, Rising, K.A, Chappell, J, Noel, J.P. | | Deposit date: | 2001-01-12 | | Release date: | 2003-06-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Single Active Site Mutations Change the Specificity of a Sesquiterpene Cyclase
To be Published
|
|
4DRQ
 
 | | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-1-((S)-1-(3,5-dichlorophenylsulfonyl)piperidine-2-carbonyloxy)-3-(3,4-dimethoxy -phenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1S)-1-[({(2S)-1-[(3,5-dichlorophenyl)sulfonyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Wang, Y, Hoogeland, B, Bracher, A, Hausch, F, Schneider, S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
1Y5W
 
 | | tRNA-guanine Transglycosylase (TGT) in complex with 6-Amino-4-[2-(4-methylphenyl)ethyl]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-[2-(4-METHYLPHENYL)ETHYL]-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
