6VK6
 
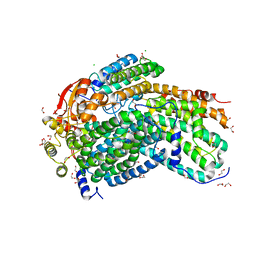 | | Crystal Structure of Methylosinus trichosporium OB3b Soluble Methane Monooxygenase Hydroxylase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Jones, J.C, Banerjee, R, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2020-01-18 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural Studies of theMethylosinus trichosporiumOB3b Soluble Methane Monooxygenase Hydroxylase and Regulatory Component Complex Reveal a Transient Substrate Tunnel.
Biochemistry, 59, 2020
|
|
6VYR
 
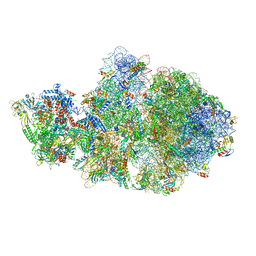 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing an 18 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
5JBH
 
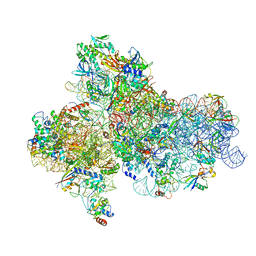 | | Cryo-EM structure of a full archaeal ribosomal translation initiation complex in the P-IN conformation | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein SX, 30S ribosomal protein eL41, ... | | Authors: | Coureux, P.-D, Schmitt, E, Mechulam, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.34 Å) | | Cite: | Cryo-EM study of start codon selection during archaeal translation initiation.
Nat Commun, 7, 2016
|
|
7DQE
 
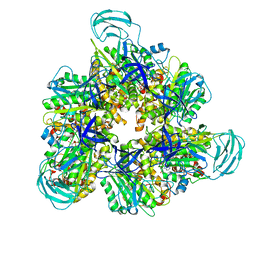 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
7ZZQ
 
 | |
8WAS
 
 | | Structure of transcribing complex 9 (TC9), the initially transcribing complex with Pol II positioned 9nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.13 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
6VZS
 
 | | Engineered TTLL6 mutant bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8WAN
 
 | | Structure of transcribing complex 4 (TC4), the initially transcribing complex with Pol II positioned 4nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.07 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
6YBH
 
 | | Deoxyribonucleoside Kinase | | Descriptor: | Deoxynucleoside kinase, GLYCEROL, Pyrrolo-dC, ... | | Authors: | Allouche-Arnon, H, Bar-Shir, A, Dym, O. | | Deposit date: | 2020-03-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computationally designed dual-color MRI reporters for noninvasive imaging of transgene expression.
Nat.Biotechnol., 40, 2022
|
|
8WAP
 
 | | Structure of transcribing complex 6 (TC6), the initially transcribing complex with Pol II positioned 6nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.85 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
7DQD
 
 | | Crystal structure of the AMP-PNP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.383 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalysis
To Be Published
|
|
8WAL
 
 | | Structure of transcribing complex 3 (TC3), the initially transcribing complex with Pol II positioned 3nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (8.52 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
6W0H
 
 | | Closed-gate KcsA soaked in 5mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
8WAO
 
 | | Structure of transcribing complex 5 (TC5), the initially transcribing complex with Pol II positioned 5nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
6W1R
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.23 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7DQC
 
 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
7DT0
 
 | | Proline hydroxylase H11-N101I mutant | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, PROLINE, ... | | Authors: | Gong, W.G, Yang, L.Y. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline
To Be Published
|
|
8WAQ
 
 | | Structure of transcribing complex 7 (TC7), the initially transcribing complex with Pol II positioned 7nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.29 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAK
 
 | | Structure of transcribing complex 2 (TC2), the initially transcribing complex with Pol II positioned 2nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.47 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAR
 
 | | Structure of transcribing complex 8 (TC8), the initially transcribing complex with Pol II positioned 8nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
7DT1
 
 | | The structure of Lactobacillus fermentum 4,6-alpha-Glucanotransferase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W.K, Yong, Y.H, Wu, L, Chen, S, Zhou, J.H, Wu, J. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43002272 Å) | | Cite: | Characterization of a new 4,6-alpha-glucanotransferase from Limosilactobacillus fermentum NCC 3057 with ability of synthesizing low molecular mass isomalto-/maltopolysaccharide
Food Biosci, 46, 2022
|
|
7ZYV
 
 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.13 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, ... | | Authors: | Sarewicz, M, Szwalec, M, Pintscher, S, Indyka, P, Rawski, M, Pietras, R, Mielecki, B, Koziej, L, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
7D66
 
 | |
7DR2
 
 | | Structure of GraFix PSI tetramer from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
7DR1
 
 | | Structure of Wild-type PSI monomer2 from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
