8KF5
 
 | |
8KF4
 
 | |
8KF3
 
 | |
8KF6
 
 | |
8KEW
 
 | |
8KF1
 
 | |
8P5F
 
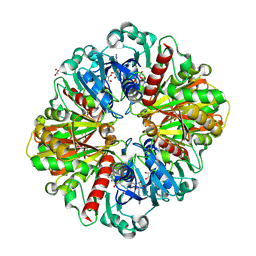 | | Human wild-type GAPDH,orthorhombic form | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Samygina, V.R, Muronetz, V.I, Schmalhausen, E.V. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S-nitrosylation and S-glutathionylation of GAPDH: Similarities, differences, and relationships.
Biochim Biophys Acta Gen Subj, 1867, 2023
|
|
1L4D
 
 | | CRYSTAL STRUCTURE OF MICROPLASMINOGEN-STREPTOKINASE ALPHA DOMAIN COMPLEX | | Descriptor: | PLASMINOGEN, STREPTOKINASE, SULFATE ION | | Authors: | Wakeham, N, Terzyan, S, Zhai, P, Loy, J.A, Tang, J, Zhang, X.C. | | Deposit date: | 2002-03-04 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of deletion of streptokinase residues 48-59 on plasminogen activation
PROTEIN ENG., 15, 2002
|
|
8P4K
 
 | | Vaccinia Virus palisade layer A10 trimer | | Descriptor: | Core protein OPG136 | | Authors: | Datler, J, Hansen, J.M, Thader, A, Schloegl, A, Hodirnau, V.V, Schur, F.K.M. | | Deposit date: | 2023-05-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Multi-modal cryo-EM reveals trimers of protein A10 to form the palisade layer in poxvirus cores.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OVB
 
 | |
8OTF
 
 | |
8PE9
 
 | | Complex between DDR1 DS-like domain and PRTH-101 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, J, Chiang, H, Xiong, W, Laurent, V, Griffiths, S.C, Duelfer, J, Deng, H, Sun, X, Yin, Y.W, Li, W, Audoly, L.P, An, Z, Schuerpf, T, Li, R, Zhang, N. | | Deposit date: | 2023-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | A highly selective humanized DDR1 mAb reverses immune exclusion by disrupting collagen fiber alignment in breast cancer.
J Immunother Cancer, 11, 2023
|
|
1AO5
 
 | | MOUSE GLANDULAR KALLIKREIN-13 (PRORENIN CONVERTING ENZYME) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLANDULAR KALLIKREIN-13 | | Authors: | Timm, D.E. | | Deposit date: | 1997-07-16 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the mouse glandular kallikrein-13 (prorenin converting enzyme)
Protein Sci., 6, 1997
|
|
2J15
 
 | | Cyclic MrIA: An exceptionally stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter. | | Descriptor: | MAI126P | | Authors: | Lovelace, E.S, Armishaw, C.J, Colgrave, M.L, Walstrom, M.E, Alewood, P.F, Daly, N.L, Craik, D.J. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-01 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Cyclic MrIA: a stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter.
J. Med. Chem., 49, 2006
|
|
1EB1
 
 | | Complex structure of human thrombin with N-methyl-arginine inhibitor | | Descriptor: | 3-CYCLOHEXYL-D-ALANYL-L-PROLYL-N~2~-METHYL-L-ARGININE, PEPTIDE INHIBITOR, THROMBIN HEAVY CHAIN, ... | | Authors: | Friedrich, R, Steinmetzer, T, Bode, W. | | Deposit date: | 2001-07-18 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Methyl Group of N(Alpha)(Me)Arg-Containing Peptides Disturbs the Active-Site Geometry of Thrombin, Impairing Efficient Cleavage
J.Mol.Biol., 316, 2002
|
|
5E7F
 
 | | Complex between lactococcal phage Tuc2009 RBP head domain and a nanobody (L06) | | Descriptor: | Major structural protein 1, nanobody L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
3MG7
 
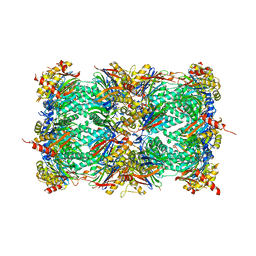 | | Structure of yeast 20S open-gate proteasome with Compound 8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(benzyloxy)-N-[(1S,2R)-2-hydroxy-1-({(1S)-1-[(2-methylbenzyl)carbamoyl]-3-phenylpropyl}carbamoyl)propyl]benzamide, MAGNESIUM ION, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
5DVH
 
 | |
3MG4
 
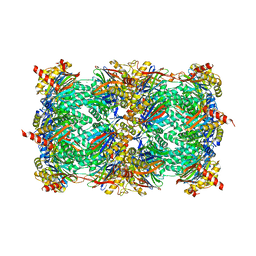 | | Structure of yeast 20S proteasome with Compound 1 | | Descriptor: | (2S)-2-amino-N-[(1S)-1-({(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}carbamoyl)-3-phenylpropyl]-4-phenylbutanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
3MG8
 
 | | Structure of yeast 20S open-gate proteasome with Compound 16 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(2,2-dimethylpropyl)-N~2~-[1H-indol-3-yl(oxo)acetyl]-L-asparaginyl-N-(2-methylbenzyl)-3-pyridin-4-yl-L-alaninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
5E3H
 
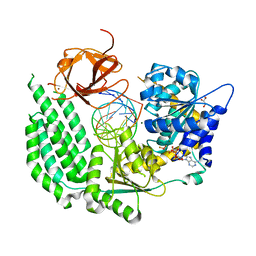 | | Structural Basis for RNA Recognition and Activation of RIG-I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Jiang, F, Miller, M.T, Marcotrigiano, J. | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of RNA recognition and activation by innate immune receptor RIG-I.
Nature, 479, 2011
|
|
5DSS
 
 | |
5DT7
 
 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group C2221 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
1ELT
 
 | |
3MG6
 
 | | Structure of yeast 20S open-gate proteasome with Compound 6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~2~-{[(1S)-6-methoxy-3-oxo-2,3-dihydro-1H-inden-1-yl]acetyl}-N-{(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}-L-threoninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
