8B5Y
 
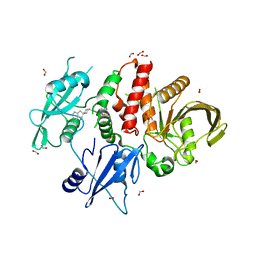 | | SHP2 in complex with allosteric imidazopyrazine inhibitor | | Descriptor: | (1~{S})-1'-[5-[2-(trifluoromethyl)pyridin-3-yl]sulfanyl-3~{H}-imidazo[4,5-b]pyrazin-2-yl]spiro[1,3-dihydroindene-2,4'-piperidine]-1-amine, FORMIC ACID, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Torrente, E, Petrocchi, A. | | Deposit date: | 2022-09-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a Novel Series of Imidazopyrazine Derivatives as Potent SHP2 Allosteric Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
1BU5
 
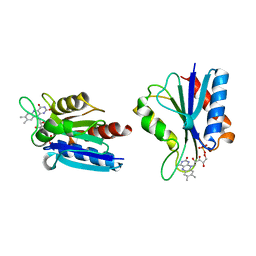 | | X-RAY CRYSTAL STRUCTURE OF THE DESULFOVIBRIO VULGARIS (HILDENBOROUGH) APOFLAVODOXIN-RIBOFLAVIN COMPLEX | | Descriptor: | PROTEIN (FLAVODOXIN), RIBOFLAVIN, SULFATE ION | | Authors: | Walsh, M.A, Mccarthy, A, O'Farrell, P.A, Mccardle, P, Cunningham, P.D, Mayhew, S.G, Higgins, T.M. | | Deposit date: | 1998-09-12 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of the Desulfovibrio vulgaris (Hildenborough) apoflavodoxin-riboflavin complex.
Eur.J.Biochem., 258, 1998
|
|
4HMX
 
 | | Crystal structure of PhzG from Burkholderia lata 383 in complex with tetrahydrophenazine-1-carboxylic acid | | Descriptor: | (1R,10aS)-1,2,10,10a-tetrahydrophenazine-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, Pyridoxamine 5'-phosphate oxidase | | Authors: | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J1N
 
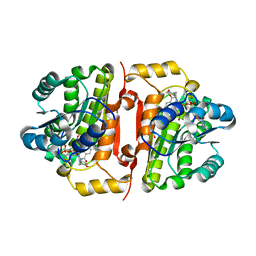 | | Crystal structures of FabI from F. tularensis in complex with novel inhibitors based on the benzimidazole scaffold | | Descriptor: | 1-(4-methoxy-3-methylbenzyl)-1,5,6,7-tetrahydroindeno[5,6-d]imidazole, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Mehboob, S, Boci, T, Brubaker, L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2013-02-01 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and biological evaluation of a novel series of benzimidazole inhibitors of Francisella tularensis enoyl-ACP reductase (FabI).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4JG9
 
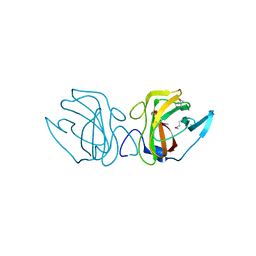 | | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis | | Descriptor: | Lipoprotein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis
To be Published
|
|
4JTQ
 
 | |
4JFB
 
 | | Crystal structure of OmpF in C2 with tNCS | | Descriptor: | Outer membrane protein F | | Authors: | Wiseman, B, Kilburg, A, Chaptal, V, Reyes-Meija, G.C, Sarwan, J, Falson, P, Jault, J.M. | | Deposit date: | 2013-02-28 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Stubborn contaminants: influence of detergents on the purity of the multidrug ABC transporter BmrA
PLoS ONE, 9, 2014
|
|
1DXY
 
 | | STRUCTURE OF D-2-HYDROXYISOCAPROATE DEHYDROGENASE | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, D-2-HYDROXYISOCAPROATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dengler, U, Niefind, K, Kiess, M, Schomburg, D. | | Deposit date: | 1996-08-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a ternary complex of D-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei, NAD+ and 2-oxoisocaproate at 1.9 A resolution.
J.Mol.Biol., 267, 1997
|
|
4J8Z
 
 | |
4IZQ
 
 | |
4J1Y
 
 | | The X-ray crystal structure of human complement protease C1s zymogen | | Descriptor: | Complement C1s subcomponent | | Authors: | Perry, A.J, Wijeyewickrema, L.C, Wilmann, P.G, Gunzburg, M.J, D'Andrea, L, Irving, J.A, Pang, S.S, Duncan, R.C, Wilce, J.A, Whisstock, J.C, Pike, R.N. | | Deposit date: | 2013-02-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6645 Å) | | Cite: | A Molecular Switch Governs the Interaction between the Human Complement Protease C1s and Its Substrate, Complement C4.
J.Biol.Chem., 288, 2013
|
|
4JKT
 
 | | Crystal structure of mouse Glutaminase C, BPTES-bound form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Fornezari, C, Ferreira, A.P.S, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Active Glutaminase C Self-assembles into a Supratetrameric Oligomer That Can Be Disrupted by an Allosteric Inhibitor.
J.Biol.Chem., 288, 2013
|
|
1EJ3
 
 | | CRYSTAL STRUCTURE OF AEQUORIN | | Descriptor: | AEQUORIN, C2-HYDROPEROXY-COELENTERAZINE | | Authors: | Head, J.F, Inouye, S, Teranishi, K, Shimomura, O. | | Deposit date: | 2000-02-29 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the photoprotein aequorin at 2.3 A resolution.
Nature, 405, 2000
|
|
4JD8
 
 | | Racemic-[Ru(phen)2(dppz)]2+] bound to synthetic DNA at high resolution | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*AP*T)-3'), Delta-Ru(phen)2(dppz) complex, Lambda-Ru(phen)2(dppz) complex, ... | | Authors: | Cook, D.S, Hall, J.P, Cardin, C.J. | | Deposit date: | 2013-02-24 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | X-ray Crystal Structure of rac-[Ru(phen)2dppz](2+) with d(ATGCAT)2 Shows Enantiomer Orientations and Water Ordering.
J.Am.Chem.Soc., 135, 2013
|
|
4JZ5
 
 | |
5W56
 
 | | Structure of Apo AztC | | Descriptor: | GLYCEROL, Periplasmic solute binding protein, SODIUM ION | | Authors: | Avalos, D, Yukl, E.T. | | Deposit date: | 2017-06-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanisms of zinc binding to the solute-binding protein AztC and transfer from the metallochaperone AztD.
J. Biol. Chem., 292, 2017
|
|
5W4H
 
 | |
8GKF
 
 | |
5W4I
 
 | |
5W4O
 
 | |
4V1L
 
 | | High resolution structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5W57
 
 | | Structure of Holo AztC | | Descriptor: | Periplasmic solute binding protein, ZINC ION | | Authors: | Avalos, D, Yukl, E.T. | | Deposit date: | 2017-06-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of zinc binding to the solute-binding protein AztC and transfer from the metallochaperone AztD.
J. Biol. Chem., 292, 2017
|
|
4UYW
 
 | |
5W4J
 
 | |
8GLO
 
 | | Haemophilus parainfluenzae Holo HphA | | Descriptor: | CHLORIDE ION, HEME B/C, Hemophilin | | Authors: | Shin, H.E, Ng, D, Moraes, T.F. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Prevalence of Slam-dependent hemophilins in Gram-negative bacteria.
J.Bacteriol., 206, 2024
|
|
