6NWL
 
 | | Structure of the Ancestral Glucocorticoid Receptor 2 ligand binding domain in complex with hydrocortisone and PGC1a coregulator fragment | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Liu, X, Ortlund, E.A. | | Deposit date: | 2019-02-06 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | First High-Resolution Crystal Structures of the Glucocorticoid Receptor Ligand-Binding Domain-Peroxisome Proliferator-ActivatedgammaCoactivator 1-alphaComplex with Endogenous and Synthetic Glucocorticoids.
Mol.Pharmacol., 96, 2019
|
|
6PUR
 
 | | Human TRPM2 bound to ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
3M9W
 
 | |
6QDV
 
 | | Human post-catalytic P complex spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Fica, S.M, Oubridge, C, Wilkinson, M.E, Newman, A.J, Nagai, K. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation.
Science, 363, 2019
|
|
1VWG
 
 | |
1NR5
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+, NAD and carbaphosphonate | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-23 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1VWR
 
 | |
5LG7
 
 | | Solution NMR structure of Tryptophan to Arginine mutant of Arkadia RING domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Birkou, M, Chasapis, C.T, Loutsidou, A.K, Bentrop, D, Lelli, M, Herrmann, T, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2016-07-06 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.
J. Mol. Biol., 429, 2017
|
|
1NVD
 
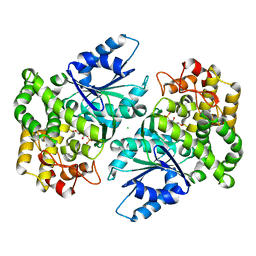 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and carbaphosphonate | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, ZINC ION, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
2OD8
 
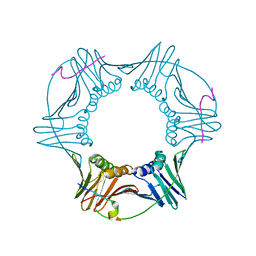 | | Structure of a peptide derived from Cdc9 bound to PCNA | | Descriptor: | DNA ligase I, mitochondrial precursor, Proliferating cell nuclear antigen | | Authors: | Chapados, B.R, Tainer, J.A. | | Deposit date: | 2006-12-21 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The C-terminal domain of yeast PCNA is required for physical and functional interactions with Cdc9 DNA ligase.
Nucleic Acids Res., 35, 2007
|
|
6PUU
 
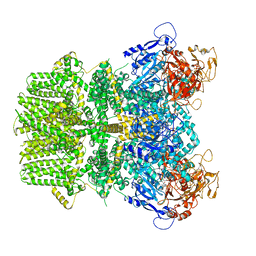 | | Human TRPM2 bound to 8-Br-cADPR and calcium | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
1VWN
 
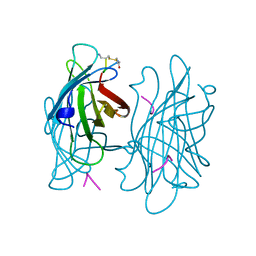 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 4.8 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
3ICU
 
 | | Protease-associated domain of the E3 ligase grail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, E3 ubiquitin-protein ligase RNF128 | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-18 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PA Domain of the E3 Ligase Grail
To be Published
|
|
1NVB
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and carbaphosphonate | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1NRX
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and NAD | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-26 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1NVA
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and ADP | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-02-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
4NEN
 
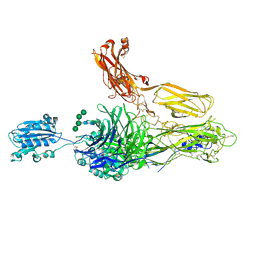 | | An internal ligand-bound, metastable state of a leukocyte integrin, aXb2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sen, M, Yuki, K, Springer, T.A. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9012 Å) | | Cite: | An internal ligand-bound, metastable state of a leukocyte integrin, alpha X beta 2.
J.Cell Biol., 203, 2013
|
|
5LG0
 
 | | Solution NMR structure of Tryptophan to Alanine mutant of Arkadia RING domain. | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Birkou, M, Chasapis, C.T, Loutsidou, A.K, Bentrop, D, Lelli, M, Herrmann, T, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2016-07-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.
J. Mol. Biol., 429, 2017
|
|
6NWK
 
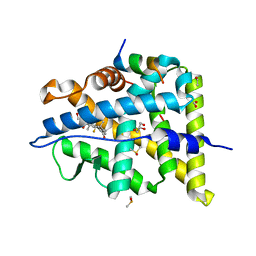 | |
6PUO
 
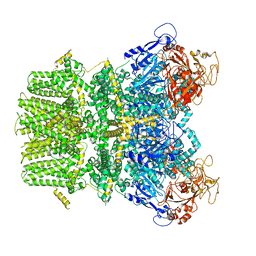 | | Human TRPM2 in the apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
4NEH
 
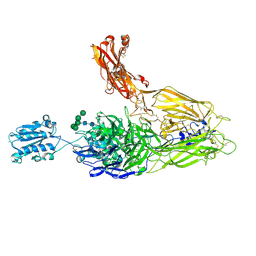 | | An internal ligand-bound, metastable state of a leukocyte integrin, aXb2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sen, M, Yuki, K, Springer, T.A. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7505 Å) | | Cite: | An internal ligand-bound, metastable state of a leukocyte integrin, alpha X beta 2.
J.Cell Biol., 203, 2013
|
|
7Y3B
 
 | | Crystal structure of TRIM7 bound to GN1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,TRIM7-GN1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1NUA
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, ZINC ION | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
8K5O
 
 | | Cryo-EM structure of the RC-LH core comples from Halorhodospira halochloris | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | Deposit date: | 2023-07-22 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural insights into the unusual core photocomplex from a triply extremophilic purple bacterium, Halorhodospira halochloris.
J Integr Plant Biol, 66, 2024
|
|
2XHS
 
 | |
